What is the full form of KBKB: Kilo Base PairKB stands for Kilo Base Pair. A base pair is a pair of two nucleotides on complementary opposing DNA or RNA strands that are joined by hydrogen bonds (often abbreviated bp). Thymine is swapped out for uracil in RNA (U). Hoogsteen base pairs are a typical example of a non-Watson-Crick base pairing with an alternative hydrogen bonding pattern, which also occurs, particularly in RNA. Base pairing is also a technique to transfer RNA anticodons and detect messenger RNA codons during protein translation. Specific base pairing patterns that designate distinct regulatory areas of genes can be recognized by some DNA- or RNA-binding enzymes. 
A base pair (bp), which consists of two nucleobases joined by hydrogen bonds, is the basic building block of double-stranded nucleic acids. Certain hydrogen bonding patterns dictate these base pairs. The genetic information stored inside each DNA strand is provided with a redundant copy thanks to the complementary nature of this base-paired structure. Because DNA is often double-stranded, the size of a single gene or an organism's whole genome is frequently measured in base pairs. Therefore, except for non-coding single-stranded sections of telomeres, the total number of base pairs equals the total number of nucleotides in one of the strands. The 23-chromosome haploid human genome is thought to be roughly 3 billion base pairs in length and comprises 20,000-25,000 unique genes. The DNA double helix's regular structure and data redundancy make it ideal for storing genetic information. DNA polymerase duplicates DNA through base pairing with incoming nucleotides, and RNA polymerase converts DNA to RNA through transcription. Numerous DNA-binding proteins can identify unique base-pairing patterns that designate distinct regulatory regions of genes. Because DNA is often double-stranded, the size of a single gene or an organism's whole genome is frequently measured in base pairs. There are 23 chromosomes in the haploid human genome, which is thought to be roughly 3.2 billion bases long and to contain between 20,000 and 25,000 different protein-coding genes. According to estimates, there are 50 billion tonnes or 5.01037 base pairs of DNA on Earth. Stability and Hydrogen bondsHigh GC content DNA is more stable than low GC content DNA. A hydrogen bond is a chemical connection underlies the base-pairing laws mentioned above. Only the "correct" pairings can form stably when hydrogen bond donors and acceptors have the proper geometrical correspondence. Contrary to popular perception, however, the hydrogen bonds do not considerably stabilize the DNA, stacking interactions are primarily responsible for stabilization. 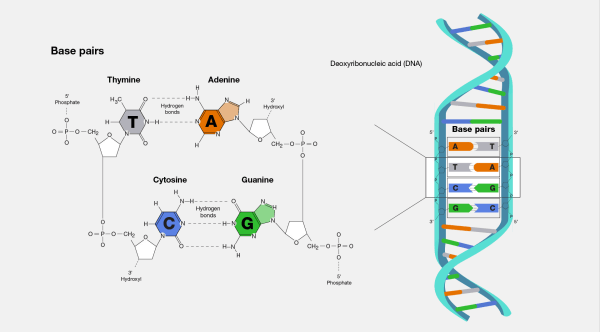
A suitable duplex structure is produced when the purine-pyrimidine base pairing of AT, GC, or UA (in RNA) occurs. However, we must match these pairings because their hydrogen acceptor and donor patterns differ. In RNA, the GU pairing with two hydrogen bonds frequently happens. At normal temperatures, paired DNA and RNA molecules are rather stable. For instance, the promoter regions for regularly transcribed genes are particularly GC-poor because they need to separate frequently. ExamplesThe DNA sequences that follow provide examples of double-stranded pair patterns. A similar RNA sequence, in which the RNA strand has uracil in place of thymine: AUCGAUUGAGCUCUAGCG UAGCUAACUCGAGAUCGC Intercalators and Base AnalogsThe incorrect use of chemical nucleotide analogs can result in non-canonical base pairing and mistakes (mainly point mutations) in DNA transcription and replication. Their isosteric chemistry is the cause. Large polyaromatic chemicals comprise most intercalators and are known or possibly carcinogenic. Examples include acridine and ethidium bromide. Repairing MismatchesErrors in DNA replication and intermediates during homologous recombination can produce mismatched base pairs. In a long sequence of healthy DNA base pairs, the mismatch repair mechanism typically needs to identify and correctly repair a minimal number of base mis pairs. To discriminate between the template strand and the freshly generated strand while repairing mismatches created during DNA replication, only the erroneous nucleotide that was just introduced is deleted (to avoid generating a mutation). The DNA mismatch repair describes the proteins involved in mismatch repair during DNA replication as well as the therapeutic implications of abnormalities in this process. 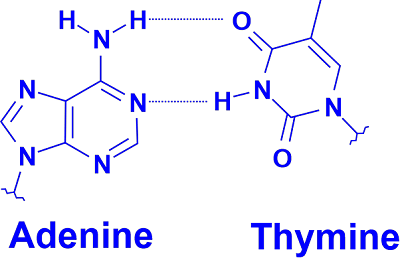
Unnatural Base Pair (UBP)An artificial base pair (UBP) is a constructed nucleobase (or DNA subunit) produced in a lab and does not exist in nature. Diverse hydrogen bonds, hydrophobic interactions, and metal coordination have all been used to support several reported new base pairs. In vitro replication of the nucleotides, which encode RNA and proteins, was effective. Following this, a high-fidelity pair in PCR amplification between Ds and 4-[3-(6-aminohexanamido)-1-propynyl]-2-tetrapyrrole (Px) was identified. They showed that the extension of the genetic alphabet considerably increases DNA aptamer affinities to the target proteins. Floyd Romesberg, a chemical biologist at the Scripps Research Institute in San Diego, California, was the leader of a team of American researchers who released findings on their research in 2012. Technically, the (d5SICS-dNaM) complex or base pair in DNA comprises these synthetic nucleotides with hydrophobic nucleobases, and two joined aromatic rings. With the help of contemporary standards in vitro techniques, such as PCR amplification of DNA and PCR-based applications, his team created various in vitro or "test tube" templates containing the unnatural base pair. It confirmed that they successfully replicated it with high fidelity in nearly all sequence contexts. Romesberg's group created it and introduced it into E. coli bacteria, which successfully replicated the synthetic base pairs over several generations. The E. coli cells ability to multiply unhindered by the transfection also showed no signs of being damaged by the organism's inborn DNA repair processes. This is the first instance of a living organism passing on a genetic code with more information to succeeding generations. To perfect the design of nucleotides that would be stable enough and reproduce as easily as the natural ones when the cells split, Romesberg claimed he and his colleagues tested 300 different iterations. Including a supporting algal gene that produces a nucleotide triphosphate transporter that effectively imports the triphosphates of d5SICSTP and dNTP into E. coli bacteria helped to achieve this in part. 
Then, a plasmid carrying d5SICS-dNaM is accurately replicated using them in the natural bacterial replication routes. The fact that the bacteria replicated these human-made DNA components shocked other experts. Successfully incorporating a third base pair would greatly increase the 20 amino acids now encoded by DNA to the theoretically feasible 172 amino acids, enhancing the possibility for living things to generate novel proteins. Although the synthetic DNA strings do not yet encode anything, scientists believe we may eventually use them to create novel proteins with potential applications in industry or medicine. According to experts, the synthetic DNA's inclusion of the unusual base pair increases the likelihood that life forms with a distinct DNA coding exist. Unorthodox Base PairingThe local backbone shape is modified along with these pairings. They have also been seen in some RNA sequences' secondary structures. Additionally, in some DNA sequences (such as the CA and TA dinucleotides), Hoogsteen base pairing-typically represented by the letters A•U/T and G•C-can coexist in dynamic equilibrium with Watson-Crick pairing. Additionally, they have been seen in a few protein-DNA complexes. Various base-base hydrogen bonds are also seen in RNA secondary and tertiary structures in addition to these alternate base pairings. These bonds are frequently required for the precise, complicated form of an RNA and its binding to interaction partners. Measurements of LengthThe length of a D/RNA molecule is frequently expressed using the following acronyms: Base pairs, or bps, are units of length along the strand that are equivalent to around 618 or 643 daltons for DNA and RNA, respectively, and 3.4 (340 pm) for one bp. Nucleotide units, abbreviated nt (or knit Mnt, or Grant) for single-stranded DNA/RNA, are employed since they are not paired. Base pairs may be designated as kbp, Mbp, GBP, etc., to distinguish them from computer storage units and bases. Although the number of base pairs it correlates to varies greatly, the centimorgan is frequently used to denote distance along a chromosome. One million base pairs make up a centimorgan in the human genome. 
As a third base pair for replication and transcription, they developed 7-(2-thienyl) imidazo [4,5-b] pyridine (Ds) and pyrrole-2-carbaldehyde (Pa) in 2006. Following this, a high-fidelity pair in PCR amplification between Ds and 4-[3-(6-aminohexanamido)-1-propynyl] -2-tetrapyrrole (Px) was identified. They showed that the extension of the genetic alphabet considerably increases DNA aptamer affinities to the target proteins. OccurrenceSingle-stranded nucleic acids can include intramolecular base pairs. Because DNA is often double-stranded, estimating the size of an organism's complete genome or a specific gene in base pairs is common. The sum of all base pairs, except for the non-coding single-stranded telomere sections, equals the number of nucleotides in one of the strands. The estimated number of DNA or base pairs on Earth is 5.01037, weighing 50 billion tonnes. Comparatively, it has been predicted that the biosphere's whole mass might reach 4 TTC (trillion tonnes of carbon). 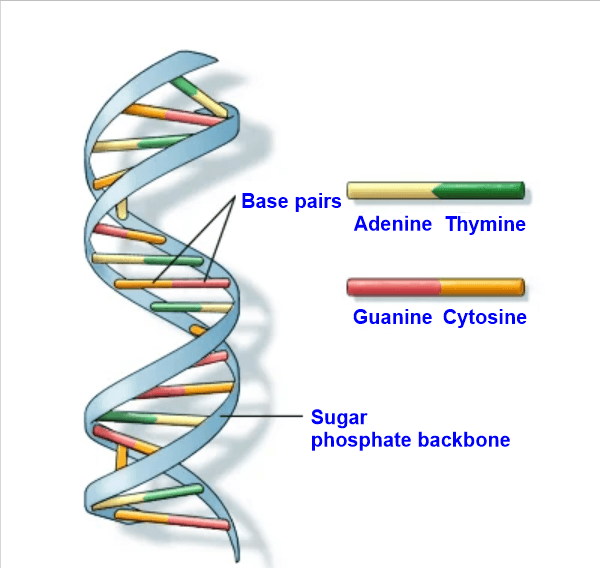
UsageBase pairs frequently estimate a gene's size within a DNA molecule. It can be challenging to detail base pairs with excessively complex genomes. Stacking basesBase stacking interactions between the pi orbitals of the aromatic rings of the bases also aid in stability, and once more, interactions with nearby bases are generally preferable. (Note, however, that a CG interaction is geometrically distinct from a GC stacking interaction with the following base pair is preferable.)
Next TopicFull Form Lists
|
 For Videos Join Our Youtube Channel: Join Now
For Videos Join Our Youtube Channel: Join Now
Feedback
- Send your Feedback to [email protected]
Help Others, Please Share










