Advantages and Disadvantages of PCRDefinition of PCRA method known as the polymerase chain reaction (PCR) is routinely used to quickly make millions to billions of copies (full or partial) of a particular DNA sample. With the help of this method, scientists can amplify a very little DNA sample (or a piece of it) to a size that is large enough to be thoroughly studied. In 1983, while working for Cetus Corporation, American biochemist Kary Mullis created PCR. The 1993 Nobel Prize in Chemistry was split between Mullis and the scientist Michael Smith, who had developed further vital methods for altering DNA. 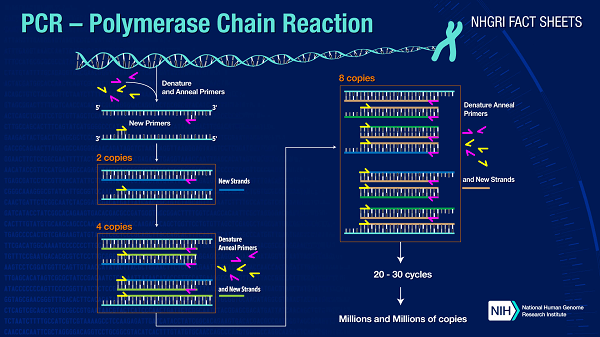
The analysis of ancient DNA samples and the detection of infectious organisms are only two examples of the many genetic testing and research techniques that heavily rely on PCR. By using PCR, which involves a series of temperature-changing cycles, extremely minute amounts of DNA sequence copies are exponentially amplified. A wide range of applications, including biological research and criminal forensics, typically need the use of PCR in medical laboratory research. PCR is essential to many of the techniques used in genetic testing and analysis, including the identification of infectious organisms and the calculation of the age of DNA samples. The use of PCR allows for the exponential amplification of copies of extremely small amounts of DNA sequences through several elemental change cycles. It is becoming more and more commonplace and frequently essential to utilize PCR in medical laboratory exams for a variety of purposes, such as biomedical research and criminal forensics. Thermal cycling is a key component of most PCR techniques. In particular, DNA melting and enzyme-driven DNA doppelganger are two temperature-dependent processes that thermal cycling imperils reactants from. An enzyme first derived from the thermophilic bacteria Thermus aquaticus called Taq polymerase is used in nearly all PCR applications. The high temperatures of the denaturation stage would cause the polymerase to denature if it was heat-sensitive. Before Taq polymerase was used, DNA polymerase had to be manually added to each cycle, which was a time-consuming and expensive operation. 
In addition to DNA cloning for sequencing, gene cloning and manipulation, gene mutagenesis, the creation of DNA-based phylogenies or functional analyses of genes, the detection of pathogens in nucleic acids, the amplification of ancient DNA, the diagnosis and monitoring of genetic disorders, gene cloning and manipulation for sequencing, gene cloning and manipulation, gene mutagenesis, gene cloning, and manipulation, gene cloning and manipulation, ancient DNA analysis tests for infectious diseases are some of the uses for the technique. In molecular biology, the polymerase chain reaction, or PCR, is a method for making several copies of a certain DNA region. Kary Mullis, a scientist from the United States, developed this plan in 1983. It has been reasonably prepared for PCR to produce millions of copies of a tiny portion of DNA. In molecular biology and biotechnology labs, this technique is used frequently. Aspects of PCR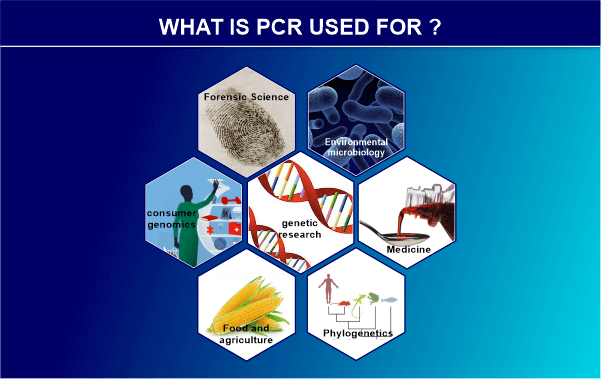
The following are PCR's constituent parts: The DNA template: The dividend from the variety's DNA. A DNA polymerase: The Taq Polymerase is used. Since it is thermostable, extremely high temperatures will not cause it to denature. Primer oligonucleotides: These are the 3' ends of the short single-stranded DNA ranges that complement the sense and antisense strings. DTTP, or Deoxyribonucleotide Triphosphate: These are the areas where DNA synthesis takes place and they also offer stability for polymerization. These are grounds that are not married. The buffer system: DNA denaturation and renaturation occur most effectively in the presence of magnesium and potassium. It is also useful for durability, polymerase mobility, and commitment. Types of PCRPCR can take one of the following forms: 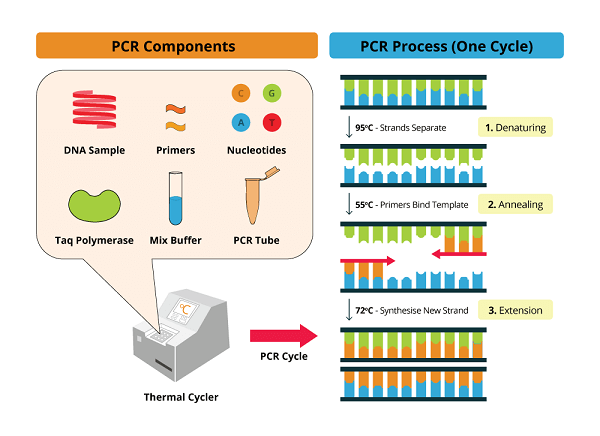
Real-time PCR: In this sample, a fluorescent writer is used to monitor DNA amplification in real time. The fluorescent reporter's signal strength immediately reflects how many amplified DNA molecules are present. Nested PCR: The goal of this was to boost sentiment and explicitness. Due to the augmentation of atypical primer binding areas, they reduce the non-specific binding of derivatives. Multi-step PCR: Using this, numerous targets can be amplified in a single PCR experiment. Multiple DNA classes are amplified at once. Quantitative PCR: This technique uses the linearity of DNA amplification to recognize, describe, and quantify an implicit classification within a category. Advantages of PCRThere are several benefits of PCR. It is fairly easy to use and comprehend, and it works quickly. A specific product can be produced in millions to billions of copies using a very sensitive approach for sequencing, cloning, and analysis. The advantages of qRT-PCR are comparable to those of PCR, but one additional advantage is the ability to quantify the outcome. Consequently, it has applications in examining changes in gene expression levels in tumors, bacteria, or other disease states. PCR is an extremely effective and useful research instrument. The PCR is used to determine the sequencing of diseases with uncertain etiologies. By using this technique, we can learn more about the disease itself and the sequence of previously undiscovered viruses that are connected to those that are currently recognized. The PCR will occupy a key position in the clinical laboratory for years to come if the process can be made even simpler and sensitive non-radiometric detection techniques can be created.
Disadvantages of PCROne significant drawback of PCR is the requirement for prior knowledge of the target sequence to create the primers that will enable its selective amplification. Accordingly, to ensure that the DNA polymerase correctly binds to the primer-template hybrids and subsequently generates the entire target region during DNA synthesis, PCR users typically need to be aware of the precise sequence(s) on each of the two single-stranded templates that are upstream of the target region. Like other enzymes, DNA polymerases can make mistakes, which causes mutations in the PCR fragments that are produced. Another drawback of PCR is that it might provide false-positive or unclear results even with minute amounts of contaminated DNA. The reagent preparation, the PCR, and the result analysis should all take place in different rooms for the investigators to reduce the possibility of contamination. Single-use aliquots of reagents should be distributed. It is recommended to regularly use pipettors with extra-long pipette tips and disposable plungers. Additionally, it is advised that a unidirectional approach be followed when setting up the lab. Never enter the PCR preparation room with any supplies or reagents used in the PCR or analysis rooms without first thoroughly decontaminating them.
The ConclusionPCR technology is used in metabarcoding investigations, the outcomes are markedly different from those of conventional, non-PCR-based sampling procedures. As a result, (1) there is no direct relationship between the number of assigned reads in an eDNA study and the abundance of the source organism, (2) there is no one-to-one correspondence between the number of assigned reads in an eDNA study and the abundance of the source organism, (3) and (4) we shouldn't expect a consistently high correlation in estimates of taxon-richness between eDNA and conventional methods. Large discrepancies in results might result from small adjustments to laboratory procedures. The usefulness of metabarcoding surveys is undeniable given that the method reveals hundreds or thousands of taxa in each sample and makes it straightforward to pinpoint biological communities among habitats and sampling locations. To make Edna a common source of data for ecological sampling, it is essential to understand the mechanisms linking amplicon reads to species' biomass or counts because many real-world applications, such as fisheries stock assessments or population surveys for endangered species, require some quantification of organisms. By concentrating on the methods by which metabarcoding data are generated, we have gained an understanding of the precise ways in which these may-and may not-be compared to other survey approaches, and in the process, we have provided a quantifiable mechanism to monitor changes in environmental samples. |
 For Videos Join Our Youtube Channel: Join Now
For Videos Join Our Youtube Channel: Join Now
Feedback
- Send your Feedback to [email protected]
Help Others, Please Share










