Dominant GeneOne allele (variant) of a gene on one copy of a chromosome can mask or override the effects of another allele (variant) of the same gene on the other copy of the chromosome, a phenomenon known as dominance. Recessive describes the second form, while dominant describes the first. This phenomenon of two separate forms of the same gene on each chromosome is caused by a mutation in one of the genes, either new (de novo) or inherited. The terms autosomal dominant and autosomal recessive are used to describe gene variants on non-sex chromosomes (autosomes) and the traits they are associated with, as opposed to those on sex chromosomes (allosomes), which have an inheritance and presentation pattern that depends on the sex of both the parent and the child. Given that the Y chromosome only has one copy, Y-linked traits cannot be dominant or recessive. Other forms of dominance include incomplete dominance, in which a gene variant only partially impacts a trait compared to when it is present on both chromosomes, and co-dominance, in which various variations on each chromosome both express their related traits. 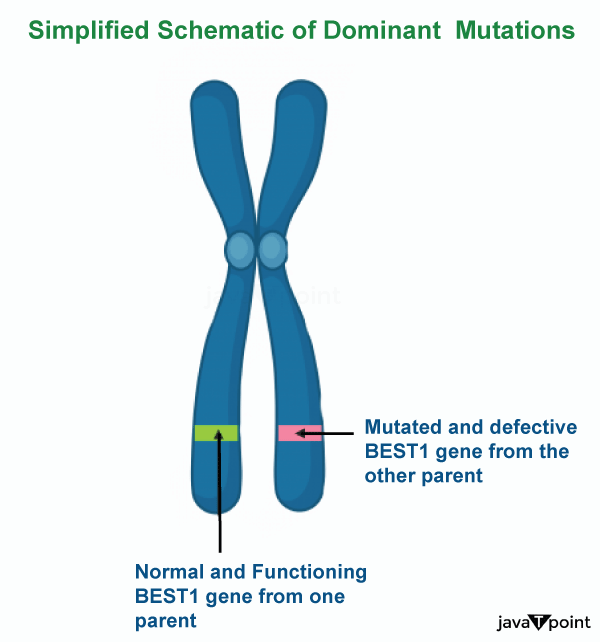
In both Mendelian inheritance and traditional genetics, dominance is a crucial idea. In order to illustrate the concepts of dominance in the classroom, letters and Punnett squares are frequently utilised. Lower-case letters are typically used for recessive alleles and upper-case letters are typically used for dominant alleles. The transmission of pea seeds is a classic case of dominance. Depending on the allele they possess, peas can either be wrinkled or round. There are three conceivable allele (genotype) pairings in this situation: RR, rr, Rr. Individuals who are RR (homozygous) produce round peas, while those who are RR (homozygous) generate wrinkled peas. Rr (heterozygous) individuals also have round peas because the R allele masks the presence of the r allele. Therefore, allele R is more prevalent than allele r, while the opposite is true for allele r. A gene's traits (phenotype) or an allele do not necessarily confer dominance. The connection between two alleles of a given gene of any function is strictly relative; one allele of a given gene may be dominant over a second allele of the same gene, recessive to a third allele of the same gene, and co-dominant with a fourth allele. Additionally, one allele could be dominant for one trait but not another. Epistasis is not the same as dominance since it happens when an allele of one gene mask the effects of an allele of another gene. Types of DominanceComplete dominanceThe effect of one allele completely masks the effect of the other in complete dominance in a heterozygous genotype. The allele that masks is dominant to the other allele, whereas the masked allele is thought to be recessive. Complete dominance in the phenotype of a heterozygote is identical to that of a dominant homozygote. The inheritance of seed shape (pea shape) in peas is a typical illustration of total dominance. Peas can be either wrinkled or spherical, depending on the allele present. There are three conceivable allele (genotype) pairings in this situation: RR, rr, Rr. Rr is heterozygous, while RR and rr are homozygotes. The RR people have peas that are wrinkly, while the RR people have peas that are round. Because the R allele hides the presence of the r allele, Rr people have round peas. As a result, allele R completely dominates allele r, and vice versa. Incomplete dominanceIncomplete dominance, also referred to as partial dominance, semi-dominance, intermediate inheritance, or, erroneously, co-dominance in the field of reptile genetics, is the term used to describe a situation in which the phenotype of the heterozygous genotype is different from and occasionally intermediate to the phenotypes of the homozygous genotypes. When heterozygous, the phenotypic outcome frequently takes the form of a melded combination of traits. For instance, either red or white is homozygous for the colour of the snapdragon flower. Pink snapdragon flowers are produced when the white homozygous bloom and the red homozygous flower are combined. Incomplete dominance led to the emergence of the pink snapdragon. When true-bred parents of white and red flowers are crossed in the four o'clock plant, pink colour results, displaying a similar type of incomplete dominance. Dominance only happens in quantitative genetics, where phenotypes are quantified and addressed numerically, when the phenotype measure of the heterozygote is closer to one homozygote than the other. A phenotype is considered to show no dominance at all if it exactly falls in the middle (numerically) of the two homozygotes. When plants from the F1 generation are self-pollinated, the phenotypic and genotypic ratio of the F2 generation will be 1:2:1 (Red:Pink:White). Co-dominanceWhen neither allele obscures the contributions of the other in the phenotype, co-dominance is present. For instance, in the ABO blood group system, three alleles?two of which are co-dominant with each other (IA, IB) and dominant over the recessive i at the ABO locus?control chemical alterations to a glycoprotein (the H antigen) on the surfaces of blood cells. Different changes are produced by the IA and IB alleles. An N-acetylgalactosamine is added to a membrane-bound H antigen by the enzyme IA. A galactose is added by the IB enzyme. There is no alteration due to the i allele. The IA and IB alleles are therefore each dominant to i, however IAIB persons have both alterations on their blood cells and hence have type AB blood, making the IA and IB alleles co-dominant. IAIA and IAi individuals both have type A blood, whilst IBIB and IBi individuals both have type B blood. Another instance is the beta-globin locus, where protein electrophoresis can be used to discriminate between the three molecular phenotypes HbA/HbA, HbA/HbS, and HbS/HbS. The alleles exhibit incomplete dominance with respect to anaemia (sickle-cell trait, a milder form distinguished from sickle-cell anaemia, is the medical condition caused by the heterozygous genotype;). Since both alleles are translated into RNA, most gene loci exhibit co-dominant expression. Incomplete dominance, when the quantitative interaction of allele products results in an intermediate phenotype, differs from co-dominance, where allelic products co-exist in the phenotypic. A red homozygous flower and a white homozygous flower, for instance, will result in offspring with red and white spots in co-dominance. The phenotypic and genotypic ratio of the F2 generation will be 1:2:1 (Red:Spotted:White) when plants of the F1 generation are self-pollinated. The ratios for incomplete dominance are the same. Once more, this traditional terminology is incorrect; in fact, such situations should not be described as exhibiting dominance at all. Addressing Common MisconceptionsThe connection between a gene's alleles is described by dominance. Typically, inheritance patterns that can be detected in Punnett Squares correspond to a dominant characteristic. If a person possesses two copies of a gene, the allele that causes the phenotype to manifest in heterozygotes is regarded as "dominant" (presuming total dominance). There are widespread myths about dominance. One typical fallacy is the idea that dominant alleles are "stronger" or "masculine" than "passive" or "feminine" recessive alleles. These beliefs are based on perceptions of dominance in non-genetic contexts, which are different from the genetic definition of dominance, such as being strong, powerful, and domineering. It is not possible to tell if an allele is beneficial, neutral, or harmful based just on dominance. However, phenotypes must be used to indirectly select genes. Dominance influences how alleles are exposed in phenotypes, which influences how quickly allele frequencies change in response to selection. Negative recessive alleles can survive in populations at low frequencies, with the majority of copies being carried by heterozygotes, with no negative effects on the heterozygotes themselves. Many hereditary genetic illnesses are caused by these uncommon recessives. NomenclatureInitially, symbols in genetics were just algebraic placeholders. The oldest convention is to capitalise the dominant allele when one allele is more common than another. The identical letter in lower case is given to the recessive allele. When the dominance relationship between the two alleles in the pea example is known, the dominant allele that results in a round form can be denoted by the capital letter R, and the recessive allele that results in a wrinkled shape by the lower-case letter r. RR, Rr, and rr, respectively, represent the homozygous dominant, heterozygous, and homozygous recessive genotypes. The three genotypes WW, Ww, and ww, the first two of which generated round peas and the third of which produced wrinkled peas, might also be used to describe the two alleles, W and w. The allele that results in the homozygous "round" or "wrinkled" phenotype is the dominant allele, regardless of whether "R" or "W" was chosen as the symbol for it. Alleles are variations of a gene. The locus symbol is followed by a different superscript for each allele. The most prevalent allele in the wild population is known as the "wild type allele" in several species. It is denoted by the superscript letter +. The wild type allele is either dominant or recessive to other alleles. The locus symbol for recessive alleles is written in lower case. The first letter of the locus symbol is capitalised for alleles that are any degree of dominance over the wild type allele. The laboratory mouse Mus musculus, for instance, has the following alleles at the a locus: Ay, dominant yellow; a+, wild type; and abt, black and tan. The Ay allele is codominant with the wild type allele while the abt allele is recessive to it. Although the Ay allele and the abt allele are codominant, demonstrating their link is outside the bounds of accepted conventions for mouse genetic nomenclature. As genetics has become increasingly sophisticated, so have the rules for genetic nomenclature. Some species' rules have been standardised by committees, but not all. Rules for one species may be slightly different from those for another species. Relationship to Other Genetic ConceptsMultiple allelesWith the exception of aneuploidies, each individual in a diploid organism only has two different alleles at any given locus; yet the majority of genes exist in many different allelic forms across the population as a whole. When the alleles affect the phenotypic differently, their dominance relationships can occasionally be characterised as a series. For instance, a number of alleles of the TYR gene, which codes for the enzyme tyrosinase, influence the coat colour of domestic cats. Different amounts of pigment are produced by the alleles C, cb, cs, and ca (full colour, Burmese, Siamese, and albino, respectively), resulting in various levels of colour dilution. The ca allele (albino) is fully recessive to the first three, while the C allele (full colour) is completely dominant over the last three. Autosomal versus sex-linked dominanceThe X and Y chromosomes are two sex chromosomes that regulate sex in humans and other mammal species. Human males are XY and females XX. The remaining chromosomal pairs are known as autosomes, and they are found in both sexes. Genetic traits linked to loci on these autosomes are referred to as autosomal, and they can be dominant or recessive. Sex-linked genetic traits are those that are linked to the X and Y chromosomes rather than those that are specific to one sex or the other. In reality, the word almost typically refers to X-linked features, and many of these abnormalities (such red-green colour blindness) are unaffected by sex. Similar to the autosomes, every gene locus on the X chromosome has two copies in females, and the same dominance relationships hold true. However, males are referred to as hemizygous for these genes since they only have one copy of each X chromosome gene locus. In comparison to the X chromosome, the Y chromosome is significantly smaller and has a much smaller collection of genes, including?but not limited to?those that affect one's "maleness," such as the SRY gene for testis-determining factor. The behaviour of sex-related gene loci in the female determines their dominance rules: since the male has only one allele (except from in rare cases of Y chromosome aneuploidy), that allele is always expressed whether it is dominant or recessive. Male birds have ZZ chromosomes and female birds have ZW chromosomes, which are the opposite of human sex chromosomes. The inheritance of qualities, however, is contrary to the XY-system; male zebra finches may possess a gene for white coloration in one of their two Z chromosomes, while females always acquire white coloration. The grasshopper XO-system exists. Males only have X, whereas females have XX. There is absolutely no Y chromosome. EpistasisEpistasis, which can occasionally mimic a dominance interaction between two separate alleles at the same locus, is an interaction between alleles at two different gene loci that affects a single attribute. The typical 9:3:3:1 ratio anticipated for two non-epistatic genes is altered by epistasis. 14 kinds of epistatic interactions are recognised for two loci. Recessive epistasis can be seen, for instance, when one gene locus dictates whether a floral colour is yellow (AA or Aa) or green (aa), while another locus decides whether the pigment is generated (BB or Bb) or not (bb). No matter if the other locus has AA, Aa, or aa genotype, a bb plant will have white flowers. The B gene exhibits recessive epistasis to the A gene rather than being dominant to the A allele because the B locus suppresses the phenotypic expression of the A locus when it is homozygous for the recessive allele (bb). This results in a typical 9:3:4 ratio in a cross between two AaBb plants, in this case yellow: flowery white: green. As in the previous illustration, where AA and Aa are yellow and aa is green, dominant epistasis allows for the determination of yellow or green pigment by a single gene locus. A second locus controls the production of a pigment precursor (dd) or not (DD or Dd). Due to the epistatic impact of the dominant D allele, the blooms of a DD or Dd plant will be colourless regardless of the genotype at the A locus. The yellow and green phenotypes are exclusively expressed in dd plants, hence in a hybrid between two AaDd plants, 3/4 of the plants will be colourless. As a result, the ratio of white, yellow, and green plants is typically 12:3:1. When two loci influence the same phenotype, supplementary epistasis happens. For instance, pigment is not created in any genotypic combination with either cc or dd if pigment colour is produced by CC or Cc but not cc, and by DD or Dd but not dd. That is, the phenotype can only be produced if both loci have at least one dominant allele. As a result, there are typically 9:7 pigmented to unpigmented plants. Contrarily, complementary epistasis only results in an unpigmented plant when the genotype is cc and dd and the ratio between pigmented and unpigmented plants is 15:1. Dominant versus AdvantageousThe terms "dominant" and "recessive" have different meanings, albeit frequently the former is mistaken with the word "advantageous" and the latter with the word "detrimental." Dominance describes the phenotype of heterozygotes in relation to the phenotypes of the homozygotes without considering how advantageous or harmful various phenotypes may be. The idea that the dominant phenotype is more superior in terms of fitness is frequently held since the word dominance has a positive connotation and because many genetic disease alleles are recessive. This is not certain, though; while the majority of genetic illness alleles are harmful and recessive, not all genetic diseases are recessive, as will be detailed later. However, this misunderstanding has persisted throughout the field of genetics' existence. One of the main reasons for the Hardy-Weinberg principle's publishing was to clear up this misunderstanding. Molecular MechanismsMendel was unaware of the chemical underpinnings of supremacy. It is now known that a gene locus is a lengthy sequence of deoxyribonucleic acid (DNA) bases or nucleotides at a specific location on a chromosome (ranging from hundreds to thousands). The fundamental tenet of molecular biology is that "DNA makes RNA makes protein"?that is, that DNA is translated into RNA to create an RNA copy, and RNA is then transcribed again to create a protein. Different alleles at a locus may or may not be transcribed in this process, and if transcribed, they may be translated into slightly different isoforms of the same protein. In the cell, proteins frequently serve as enzymes that catalyse chemical events that either directly or indirectly result in phenotypes. Genome-wide mutations can change catalytic activity, which has an impact on dominance. The DNA sequences of the two alleles present at any gene locus in a diploid organism may be the same (homozygous) or different (heterozygous). The proteins produced by each allele may be identical even though the gene locus is heterozygous at the DNA sequence level. Both alleles cannot be deemed dominant when there is no distinction between the protein products (see co-dominance, above). Again, neither allele can be claimed to be dominant since even if the two protein products (allozymes) are slightly different, it is likely that they create the same phenotype with regard to enzyme action. ZygosityAlleles are thought to sort independently, with one allele being "dominant," according to Mendel's Law of Independent Assortment. Dominance may be impacted by zygosity, or the degree of similarity between an organism's alleles. These would be determined by the allele-haplotype interactions inside a diploid organism. Due to gene haploidy, a single functioning allele may produce enough protein to produce a phenotype that is identical to the homozygote. There are three different types of potential haplotype interactions: 1. HaplosufficiencyA haplosufficient gene's functional allele in a diploid organism would be regarded as dominant, whilst an inactive allele would be regarded as recessive. Assume, for instance, that the functional homozygote produces 100% of the normal amount of enzyme, with 50% coming from each of the two functional alleles. The heterozygote's one functioning allele generates 50% of the necessary amounts of enzyme, which is enough to result in the expected phenotype. The functional allele is dominant over the non-functional allele if the phenotypes of the heterozygote and functional-allele homozygote are the same. At the albino gene locus, this happens: the heterozygote produces an adequate amount of the enzyme needed to transform the pigment precursor into melanin, and the person has normal pigmentation. For instance, the albino phenotype of humans and other creatures arises when a person is homozygous for an allele that codes for a non-functional version of an enzyme required to create the skin pigment melanin. 2. Incomplete HaploinsufficiencyA phenotype that is not normal but is less severe than that of the non-functional homozygote is less frequently produced by the presence of a single functional allele. The words haplo-insufficiency and incomplete dominance are frequently used to describe these situations because this occurs when the functional allele is not haplo-sufficient. The intermediate interaction occurs when a phenotype intermediate between the two homozygotes is produced by the heterozygous genotype. One allele is considered to exhibit incomplete dominance over the other depending on which of the two homozygotes the heterozygote most closely resembles. For instance, the Beta-chain protein (HbB), one of the two globin proteins that make up the blood pigment haemoglobin, is produced in humans by the Hb gene locus. Many people carry the HbA allele homozygously, but other people are heterozygous or homozygous for the HbS alternative allele. HbS/HbS homozygotes experience a shift in haemoglobin molecule shape that alters the morphology of red blood cells and results in sickle-cell anaemia, a severe and potentially fatal form of anaemia. People who are heterozygous HbA/HbS for this allele experience sickle-cell trait, a considerably less severe form of anaemia. The HbA allele is said to be incompletely dominant to the HbS allele because the illness phenotype of HbA/HbS heterozygotes is more comparable to but not exactly the same as the HbA/HbA homozygote. 3. Complete HaploinsufficiencyOne functional allele in a heterozygote may not produce enough gene product for all of the gene's functions, leading to the dominance of the typically non-functional alleles. Instead of the wild type, the phenotype will thus resemble that of a homozygote with a non-functional allele. It would be correct to say that the functional allele of the wild-type phenotype is dominant over the non-functional allele. This circumstance could arise if the protein generated by the non-functional allele interferes with the regular allele's protein's ability to function properly. The heterozygote's illness phenotype more closely matches that of the homozygote for two defective alleles because the defective protein "dominates" the normal protein. It is common practise to wrongly refer to defective alleles that result in a different phenotype when heterozygous with the normal allele as "dominant" even when their homozygous phenotype has not been studied. Numerous trinucleotide repeat disorders, such as Huntington's disease, exhibit this characteristic. Complete haploinsufficiency results in the mutant protein's dominant action in Huntington's disease. The HTT gene normally has about 20 C-A-G nucleotide repeats, however individuals with Huntington Disease have 40 or more C-A-G repeats. Another illustration is the connective tissue disorder Marfan syndrome, which is brought on by a mutation in the fibrillin-1 (FBN1) gene. One parent passes down a dominant aberrant FBN1 gene copy, whereas the other parent passes down a normal copy of the FBN1 gene. Dominant-Negative MutationsMany proteins are often active as multimers, also referred to as homomultimeric proteins or homooligomeric proteins, which are collections of numerous copies of the same protein. In reality, the BRENDA Enzyme Database has 83,000 unique enzymes from 9800 unique species, and the majority of these constitute homooligomers. A mixed multimer can form when both the wild-type and mutant forms of the protein are present. A dominant-negative mutation is one that creates a mutant protein that interferes with the function of the wild-type protein in the multimer. Human somatic cells are susceptible to dominant-negative mutations that give the mutant cell a proliferative advantage and cause it to grow clonally. For instance, a cell may become resistant to apoptosis if a dominant-negative mutation occurs in a gene required for the normal process of programmed cell death (Apoptosis) in response to DNA damage. This will enable clone growth even in the presence of significant DNA damage. The tumour suppressor gene p53 carries these dominant-negative mutations. The P53 wild-type protein often exists as an oligotetramer, which is a four-protein multimer. Many various cancers and pre-cancerous lesions (such as brain tumours, breast cancer, oral pre-cancerous lesions, and oral cancer) have dominant-negative p53 mutations. There are dominant-negative mutations in additional tumour suppressor genes as well. For example, the Ataxia Telangiectasia Mutated (ATM) gene, which increases susceptibility to breast cancer, has two dominant-negative germ line mutations. Acute myeloid leukaemia can be brought on by dominant-negative mutations of the transcription factor C/EBP. Dominant negative mutations that are inherited can raise the risk of illnesses outside cancer. Peroxisome proliferator-activated receptor gamma (PPAR) dominant-negative mutations are linked to severe insulin resistance, diabetes mellitus, and hypertension. Other than humans, other creatures have also been described with dominant-negative mutations. In fact, the bacteriophage T4 tail fibre protein GP37 was the subject of the first investigation to document a mutant protein limiting a wild-type protein's ability to perform normally in a mixed multimer. In investigations involving P53, ATM, C/EBP, and bacteriophage T4 GP37, mutations that result in shortened mutant proteins rather than full-length mutant proteins appear to have the most dominant-negative effect. Dominant and Recessive Genetics in HumansMendelian diseases are often influenced by dominance, while complex features are typically controlled by additive alleles. Numerous genetic disorders or features in humans are categorised as "dominant" or "recessive" by default. Particularly when it comes to so-called recessive disorders, which are in fact caused by recessive genes but might oversimplify the underlying biological foundation and result in a misperception of the nature of dominance. For instance, any of the several ( 60) alleles at the gene locus for the enzyme phenylalanine hydroxylase (PAH) might cause the recessive genetic illness phenylketonuria (PKU). The substrate phenylalanine (Phe) and its metabolic byproducts accumulate in the central nervous system as a result of many of these alleles producing little to no PAH, which can lead to severe intellectual impairment if left untreated. The quantity of phenylalanine in the blood is around 60 M (= mol/L) in unaffected individuals homozygous for a standard functional allele (AA), and PAH activity is standard (100%). Untreated people who are homozygous for one of the PKU alleles (BB) exhibit PKU and have PAH activity that is near to zero and Phe that is thirty to forty times normal. Blood Phe is raised twofold, PAH activity is only 30% (rather than 50%) of normal, and PKU is not present in the AB heterozygote. Therefore, the B allele is only partially dominant to the A allele with respect to its molecular function, which is the determination of PAH activity level (0.3% 30% 100%), whereas the A allele is dominant to the B allele with respect to PKU. As of 60 M, 120 M, and 600 M, the A allele is only partially dominant to the B allele with respect to Phe. Note once more that the fact that the recessive allele results in a more extreme Phe phenotype is unimportant to the question of dominance. A CC homozygote for the third allele C generates very little PAH enzyme, which leads to hyperphenylalaninemia, a disorder that does not cause intellectual disability but causes a slightly increased level of Phe in the blood. Because of this, the dominance relationships of any two alleles may change depending on whatever phenotypic feature is being examined. Instead of trying to shoehorn genotypes into dominant and recessive categories, it is usually more instructive to discuss the phenotypic effects of the allelic interactions involved.
Next TopicGene Library
|
 For Videos Join Our Youtube Channel: Join Now
For Videos Join Our Youtube Channel: Join Now
Feedback
- Send your Feedback to [email protected]
Help Others, Please Share









