Gene Regulation in ProkaryotesProkaryotic cells' cytoplasmic nucleoid contains a circular chromosome that is organized into a supercoiled circular structure by its DNA. Blocks of DNA, known as operons, are used to encode proteins that are required for a particular function. In the lactose (or lac) operon, for instance, all the genes required to utilize lactose as a fuel are located close to one another. Three classes of regulatory molecules?repressors, activators, and inducers?can influence the expression of operons in prokaryotic cells. In contrast to activators, which boost gene transcription in response to an external stimulus, repressors are proteins that inhibit a gene's transcription. Last but not least, inducers are tiny chemicals that activate or repress transcription based on cellular requirements and substrate availability. In bacteria and archaea, structural proteins with related functions?such as the genes for the enzymes that catalyse the various steps in a single biochemical pathway?are typically encoded together within the genome in a block known as an operon and are cotranscribed under the direction of a single promoter. As a consequence, the polycistronic transcript is produced. Since they will either be needed all at once or not at all, the promoter has simultaneous control over the regulation of these structural genes' transcription. Researchers from the Pasteur Institute in France, Fran�ois Jacob (1920-2013), and Jacques Monod, were the first to show how bacterial genes are arranged into operons via their study of the lac operon of E. coli. They found the sole promoter for the lac operon, which includes all of the structural genes encoding the enzymes needed to use lactose as an energy source, in E. coli is the lac promoter. In recognition of their work, they won the Nobel Prize in Physiology or Medicine in 1965. DNA sequences that regulate an operon's transcription may be located in a region called the regulatory region and are present in every operon. The regulatory region, which includes the promoter and its surrounds, is where transcription factors, which are proteins encoded by regulatory genes, may bind. In order to transcribe structural genes, RNA polymerase must first bind to the promoter. Transcription factors influence this process. A transcription factor known as a repressor attaches to a DNA sequence within the operator regulatory region, which is located between the promoter's RNA polymerase binding site and the first structural gene's transcriptional start site, to prevent the transcription of a gene in response to an external stimulus. This operator regulatory region is involved in regulating gene expression. Physically, RNA polymerase cannot transcribe structural genes while repressors are bound. By making it simpler for RNA polymerase to bind to the promoter, an activator, on the other hand, is a transcription factor that increases a gene's transcription in response to an external stimulus. The third class of regulatory molecules, called inducers, are very small molecules that engage with activators or repressors to either promote or stop transcription. In prokaryotes, there exist instances of operons whose gene products are often needed but whose expression is uncontrolled. These operons are constitutively expressed, which implies that they are continuously translated and transcribed to provide the cell stable intermediate levels of the protein products. These genes produce the essential metabolic enzymes as well as those involved in cellular maintenance tasks like DNA replication, repair, and expression. On the other hand, other bacterial operons are only expressed when necessary and are controlled by repressors, activators, and inducers. Regulation of Gene Expression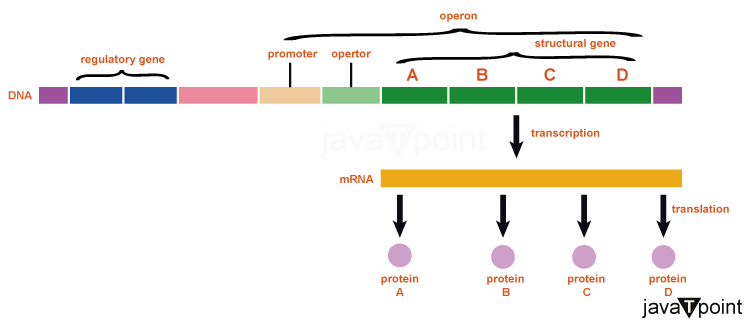
For a cell to function properly, the proper amount of time must be allowed for the creation of necessary proteins. Not all DNA-encoded genes in a certain cell type are translated into RNA or transcribed into protein as a result of the distinctive functions of diverse cells in our body. The specialised proteins that make up the eye (iris, lens, and cornea), in contrast to the specialised proteins in the heart (pacemaker cells, heart muscle, and valves), are exclusively expressed in the eye. Gene expression is the process of activating a gene to create RNA and protein. Each cell regulates the timing and manner of gene expression, whether the organism is a simple unicellular one or a sophisticated multicellular one. An intricate regulatory system governs how a gene is expressed. In single-celled creatures like prokaryotes, it makes sure that a cell's resources aren't squandered on proteins that the cell doesn't require at that particular moment. In multicellular species, it allows for cellular differentiation. It is crucial to our knowledge of human health to clarify the processes governing gene expression. Cancer and other illnesses emerge in people when this mechanism is dysfunctional. It is crucial for comprehending a specific infectious illness to comprehend the interplay between a pathogen's gene expression and that of its human host. OperonBacterial genes that perform similar tasks, such as those that code for the enzymes that catalyze the several stages in a single metabolic process, are controlled collectively and located close to one another on the DNA. This group of genes shares a regulatory region that regulates the transcription of the unit and ONE promoter. A collection of these genes is known as an OPERON. The OPERON is converted into a polycistronic mRNA, a single mRNA that contains the information required to make several proteins. The promoter concurrently regulates the regulation of the transcription of these structural genes, which will either be needed all at once or none at all. The ability of bacteria to quickly adapt to environmental changes is enabled by the grouping of similar genes under a single regulatory mechanism. The trp Operon: A Repressor Operon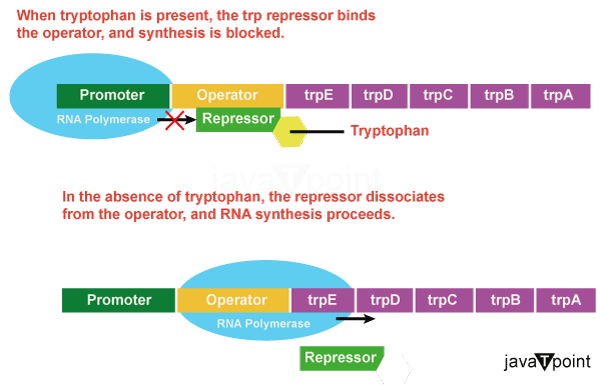
Bacteria such as E. coli cannot survive without amino acids. Tryptophan is one of the amino acids that E. coli may consume from the environment. E. coli has the ability to synthesise tryptophan utilising enzymes that are expressed by five distinct genes. All five of these genes are part of the tryptophan operon (trp). If tryptophan is already available in the environment, E. coli does not have to synthesise it, therefore the switch that governs the activation of the genes in the trp operon is disabled. The switch governing the operon, transcription, gene expression, and tryptophan synthesis are all activated when the tryptophan supply is low. The promoter, operator, and five genes called trpE, trpD, trpC, trpB, and trpA are all found in the trp operon and are arranged on the DNA in a particular order. The promoter is bound by RNA polymerase. RNA synthesis is inhibited when tryptophan is present because the trp repressor attaches to the operator and inhibits the RNA polymerase from passing the operator. The repressor separates from the operator in the absence of tryptophan. Now that the operator can slide aside, transcription can start. Within the trp operon, there are five genes that are required for E. coli to produce tryptophan. When the amino acid is in abundant, two tryptophan molecules bind to the repressor protein at the operator sequence. As a result, the RNA polymerase cannot physically transcribe the tryptophan genes. Tryptophan is required for the repressor protein to link to the operator, which prevents transcription of the genes in the absence of tryptophan. The coding area refers to a DNA sequence that codes for proteins. On the chromosome, the operon contains the five coding areas for the tryptophan biosynthetic enzymes. The transcriptional start site is the area right before the coding region. When RNA polymerase binds to this area of DNA, transcription can begin. Each operon has a region within or close to the promoter to which proteins (activators or repressors) can bind and regulate transcription. The promoter sequence is upstream of the transcriptional start point. Between the promoter area and the first trp coding gene, there is a DNA sequence called the operator sequence that is encoded. The DNA code for this operator is present, and the repressor protein can bind to it. Two tryptophan molecules attach to the trp repressor when tryptophan is present in the cell, causing it to alter its shape and bind to the trp operator. When the tryptophan-repressor complex binds to the operator, RNA polymerase cannot bind and cannot transcribe the downstream genes. The operon is active, and tryptophan is synthesized when tryptophan is absent from the cell because the repressor does not bind to the operator on its own. The trp operon is negatively regulated, and the proteins that bind to the operator to mute trp expression are known as negative regulators. This is because the repressor protein actively attaches to the operator to keep the genes turned off. Within the trp operon, there are five genes that are required for E. coli to produce tryptophan. Two tryptophan molecules bind to the repressor protein at the operator sequence when there is an abundance of the amino acid. This physically prevents the tryptophan genes from being transcribed by the RNA polymerase. In the absence of tryptophan, the repressor protein fails to connect to the operator, allowing the transcription of the genes. Catabolite Activator Protein (CAP): An Activator Regulator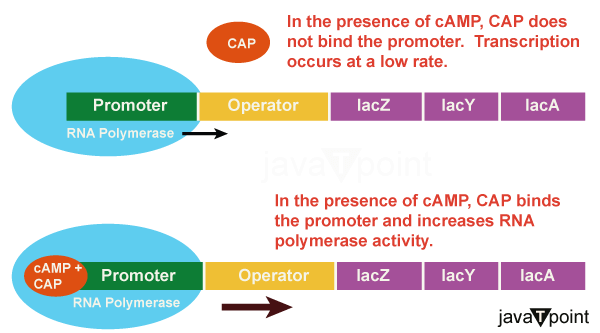
Proteins that bind to operator sequences function as positive regulators, activating and turning on genes, just how tryptophan molecules negatively control the trp operon. For instance, when there is a lack of glucose, E. coli bacteria may utilise other sugar sources. To do this, it is required to transcribe new genes to process these alternative genes. As blood glucose levels drop, cyclic AMP (cAMP) starts to accumulate in the cell. In E. coli, the signalling molecule cAMP controls the metabolism of glucose and energy. When the cell's glucose levels drop, the positive regulator catabolite activator protein (CAP), a protein that binds to the promoters of operons that control the metabolism of alternative sugars, is activated. Then, accumulating cAMP adheres to CAP. The complex that is created when cAMP and CAP interact binds to the promoter region of the genes necessary for using alternate sugar sources. The promoter of these operons has a CAP binding site upstream of an RNA polymerase binding site. This enhances the genes' capacity to be expressed as well as RNA polymerase's capacity to attach to the promoter region. When glucose levels fall, E. coli may use other sugars as fuel, although doing so requires the transcription of additional genes. As glucose sources become limited, cAMP levels increase. An operator region upstream of the genes that need to use alternate sugar sources is bound to by the positive regulator CAP protein and thus cAMP. The Lac Operon: An Inducer Operon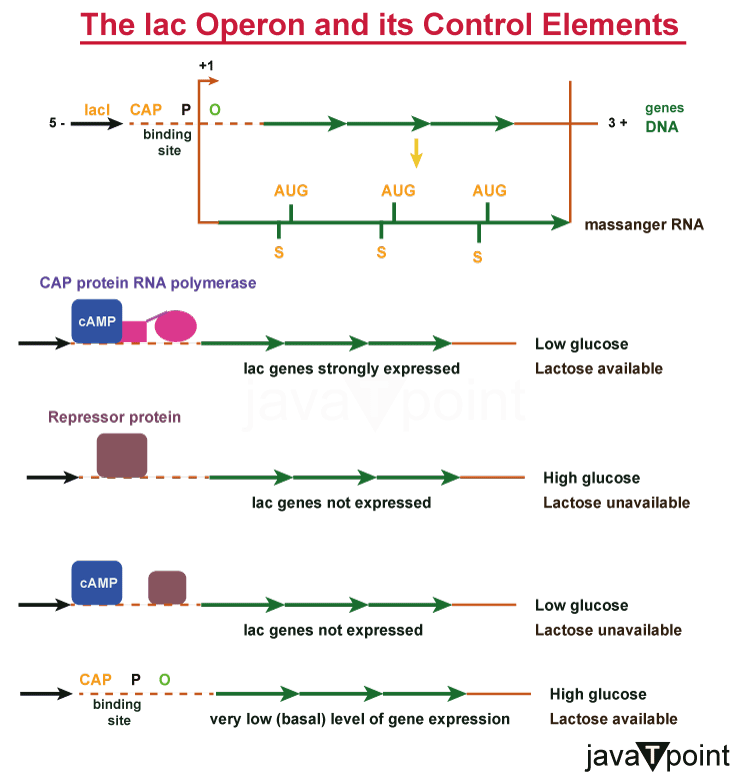
The lactose operon (lac operon) is a crucial operon for the transport and metabolism of lactose in many different enteric bacteria, including E. coli. The lac operon permits effective lactose digestion in the case that glucose is not easily available, even though most bacteria choose glucose as their preferred carbon source. The action of beta-galactosidase makes this feasible. Being the first well studied genetic regulatory system, the lac operon's gene control is a notable example of prokaryotic gene regulation. For this reason, it is often discussed in introductory courses on molecular and cellular biology. Fran�ois Jacob and Jacques Monod examined the lactose metabolism system to learn how a biological cell determines which enzyme to synthesise. For their study of the lac operon, they were awarded the Nobel Prize in Physiology in 1965. Bacterial operons are multi-protein-producing polycistronic transcripts that may be made from a single mRNA transcript. In this instance, the three genes of the lac operon?lacZ, lacY, and lacA?can be expressed, and their ensuing proteins are translated when the bacteria need lactose as a source of sugar. Lactose, a disaccharide, is broken down into glucose and galactose by the enzyme -galactosidase, which is the gene product of lacZ. Beta-galactoside permease, a membrane protein that integrates into the cytoplasmic membrane to allow lactose to enter the cell, is encoded by lacY. Last but not least, lace produces -galactoside transacetylase. Producing enzymes in the absence of lactose or in the presence of a more advantageous energy source, such as glucose, would be wasteful. The lac operon employs a two-part regulation system to make sure that the cell only utilizes energy to produce the enzymes it has been programmed to encode. The lac repressor lacI prevents the lac operon's enzymes from being produced in the absence of lactose. Unless a co-inducer attaches to it, the lac repressor is always produced. In other words, transcription only occurs when modest co-inducer molecules are present. EIIAGlc inhibits lactose permease to prevent the transport of lactose into the cell, and the catabolite activator protein (CAP), required for the synthesis of the enzymes, is inactive in the presence of glucose. This dual regulatory mechanism is what leads to diauxic, which is the sequential utilisation of glucose and lactose in two distinct developmental phases.
Next TopicSupplementary Gene
|
 For Videos Join Our Youtube Channel: Join Now
For Videos Join Our Youtube Channel: Join Now
Feedback
- Send your Feedback to [email protected]
Help Others, Please Share









