Gene Regulation in EukaryotesThe human genome is thought to include roughly 25,000 genes, about the same amount as maize and about twice as many as common fruit flies. Even more remarkable is the fact that 1.5% of the genome, or around 25,000 genes, are encoded. What precisely does the remaining 98.5% of our DNA accomplish, then? While there are still many unanswered questions about the purpose of the additional sequence, we do know that it does include sophisticated instructions that govern the complicated on-and-off switching of gene transcription. RNA polymerase binds to the gene's promoter to initiate transcription, which is a fundamental similarity between prokaryotes and eukaryotes. However, multicellular eukaryotes control cell differentiation through more complex and precise temporal and spatial regulation of gene expression. Prokaryotes have a significantly smaller genome than multicellular eukaryotes, which are arranged into numerous chromosomes with more complicated sequences. Genes having the same sequences are found in many eukaryotic species of plants and animals. Additionally, all diploid, nucleated cells in an organism contain the same DNA sequences (but not the same proteins), despite the fact that these cells give rise to tissues with drastically different appearances, traits, and functions. So why is there such a wide range of variation between and within these organisms? Simply put, the variation we perceive in nature is produced by the way certain genes are switched on and off in particular cells. In other words, specific gene regulation results in the production of distinct functionalities in various cell types. Naturally, higher eukaryotes continue to regulate their genes in response to external signals. However, interactions between cells inside the organism that direct development result in a further layer of control. In particular, there are two degrees of control over gene expression. The first way transcription is regulated is by regulating the quantity of mRNA that can be generated from a given gene. The second level of regulation, post-transcriptional mechanisms, control how mRNA is translated into proteins. After a protein is generated, post-translational modifications may still have an effect on how it performs. Eukaryotic Epigenetic Gene RegulationOver 20,000 genes are encoded by the human genome, and each of the 23 pairs of chromosomes contains hundreds of genes. In order to fit within the nucleus, the DNA is properly wrapped, folded, and compressed into chromosomes. Furthermore, it is set up such that each sort of cell may access a certain section as required. The wrapping of DNA strands around histone proteins is the first stage of organization or packing. Histones bundle and arrange DNA into nucleosome complexes, which are structural elements that may regulate how proteins can access certain DNA areas. When seen under an electron microscope, the way that DNA is wound around histone proteins to produce nucleosomes resembles a string of tiny beads. These beads (histone proteins) may travel along the DNA string, altering the molecule's structure. 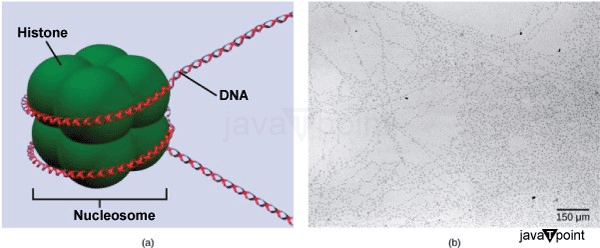
If transcription of that gene is to occur, the nucleosomes around the area of DNA that codes for that gene may travel along the DNA to open that specific chromosomal region and allow the transcriptional machinery (RNA polymerase) to begin transcription. Nucleosomes migrating to open the chromosomal structure may disclose a length of DNA, but only under very close observation. Epigenetic regulation is a term used to describe this kind of gene control. The term "epigenetics" means "around genetics." The DNA and histone proteins are temporarily altered, while the nucleotide sequence is unaltered. The structure of the chromosomes (open or closed) is changed as needed, although these changes are only temporary (although they often persist through several rounds of cell division). A gene may be activated or inactivated based on the location and modifications made to the DNA and histone proteins. Eukaryotic Transcription Gene RegulationSimilar to prokaryotic creatures, RNA polymerase in eukaryotes binds to a sequence upstream of a gene to begin transcription of that gene. Contrary to prokaryotic organisms, eukaryotic RNA polymerase needs other proteins, sometimes known as transcription factors, to help in transcription start. Proteins known as transcription factors bind to regulatory regions such as the target gene's promoter and other sequences to regulate transcription. Transcription in eukaryotic cells cannot be started by RNA polymerase alone. To attract RNA polymerase to the transcription site, transcription factors must first bind to the promoter region. The Promoter and the Transcription MachineryA DNA segment known as an enhancer encourages transcription. Distal control elements are little DNA sequences that are the building blocks of every enhancer. Mediating proteins and transcription factors engage in interaction with activators attached to the distal control elements. Two distinct genes can share a promoter yet have distinct distal regulatory elements, allowing for divergent gene expression. To make controlling gene expression simpler, genes are organized. The coding sequence is located just before the promoter region. The promoter's job is to bind transcription factors that regulate how transcription starts. Some areas aid in enhancing or increasing transcription in various eukaryotic genes. These areas, which are known as enhancers, are only sometimes near the genes that they improve. A gene's coding region, downstream of a gene, upstream of a gene, or hundreds of nucleotides distant are all possible locations for them. The locations or binding sequences for enhancer regions are transcription factor binding sequences. The form of the DNA changes when a DNA-bending protein attaches to it. This shapeshift enables the interaction between the activators linked to the enhancers and the RNA polymerase, as well as the transcription factors bound to the promoter region. Regulation of gene expression is made easier by the way genes are structured. The promoter region is immediately upstream of the coding sequence. This area may be only a few nucleotides long or hundreds of nucleotides long. Longer promoters provide greater surface area for proteins to bind to. This also provides the transcription process more authority. The size of the promoter varies widely across genes and depends on the particular gene. As a consequence, there may be significant differences in how much each gene is regulated. The promoter's role is to bind transcription factors that control how transcription begins. An enhancer located far upstream and a promoter located right next to the gene regulate the expression of eukaryotic genes. The enhancer and promoter are near each other after the DNA folds over itself; mediator proteins and transcription factors lie between the enhancer and the promoter. Distal control elements are short DNA sequences found inside the enhancer that bind activators, which in turn bind mediator proteins and transcription factors attached to the promoter. The complex is bound by RNA polymerase, which starts transcription. Enhancers on various genes have various distal control elements, which enable varied regulation of transcription. A DNA sequence that encourages transcription is known as an enhancer. Distal control elements are little DNA sequences that make up each enhancer. Transcriptional factors and mediator proteins communicate with activators linked to distal regulatory elements. Differential gene expression is made possible by the possibility that two distinct genes share a promoter but have separate distal regulatory elements. In the promoter region, just upstream of the transcriptional start point, is where the TATA box is found. In this box, there are just thymine and adenine dinucleotides, or TATA repeats. RNA polymerase binds to the transcription initiation complex to make transcription possible. The TATA box must first be bound by a transcription factor (TFIID) in order for transcription to begin. The TATA box is drawn to the TATA box by TFIID binding, which also attracts the other transcription factors TFIIB, TFIIE, TFIIF, and TFIIH. After assembling this complex, RNA polymerase may bind to the upstream sequence. When it is joined with the transcription factors, RNA polymerase is phosphorylated. This results in the activation of the transcription initiation complex and the proper positioning of the RNA polymerase for transcription to begin. Aside from interacting with transcription factors and mediation proteins, DNA-bending proteins also bring enhancers-which may be dispersed across the genome-into touch. Other transcription factors may bind to the promoter in addition to the typical transcription factors to control gene transcription. The promoters of a certain set of genes are where these transcription factors bind. They are attracted to a particular sequence on the promoter of a particular gene rather than being universal transcription factors that bind to all promoter complexes. A cell has hundreds of transcription factors, and each one of them has a unique DNA sequence motif that it may bind to. The promoter directly upstream of the encoded gene is referred to as a cis-acting element when transcription factors bind to it since it is located on the same chromosome next to the gene. The transcription factor binding site is the area at which a certain transcription factor binds. The proteins locate their binding sites and begin the transcription of the required gene as a result of transcription factors' reactions to environmental cues. Enhancer and Transcription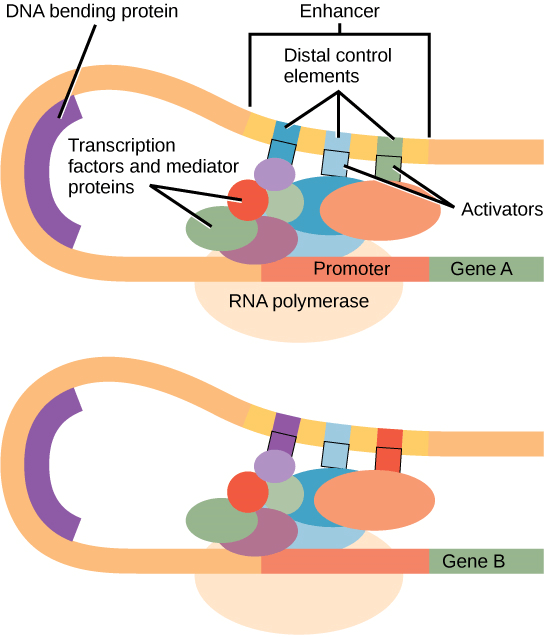
Some eukaryotic genes contain sequences that aid in enhancing or increasing transcription. These areas, known as enhancers, are only sometimes near the genes they boost. They might be thousands of nucleotides apart, upstream of the gene, within the gene's coding region, or downstream of the gene. Enhancer regions are the locations where transcription factors bind. A DNA-bending protein binds, altering the form of the DNA. This change in form allows for interaction between the activators connected to the enhancers and the RNA polymerase, as well as the transcription factors belonging to the promoter region. DNA is a three-dimensional entity, despite the fact that it is commonly visualised as a two-dimensional, straight line. A nucleotide sequence may fold over and interact with a certain promoter even if it is thousands of nucleotides distant from another. Eukaryotic cells contain ways to stop transcription, just like prokaryotic cells do. Repressors of transcription can attach to enhancer or promoter regions and stop transcription. Repressors react to environmental cues similarly to transcriptional activators in order to block the binding of activating transcription factors. Post Translational Control of Gene Expression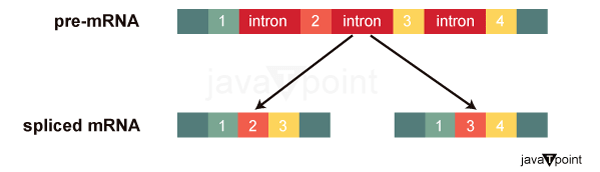
After RNA has been digested to a mature condition, translation may begin. The processing that occurs after an RNA molecule has been translated but before it is translated into a protein is referred to as post-transcriptional modification. Similar to the transcriptional and epigenetic phases of processing, the post-transcriptional step of processing may also be modified to control gene expression in the cell. Only if the RNA is processed, shuttled, or translated can protein synthesis occur. RNA SplicingThe RNA transcript in eukaryotic cells frequently contains introns, which are cut off before translation. RNA's exons are responsible for encoding proteins. After a molecule of RNA has been transcribed but before it exits the nucleus to be translated, it is processed, and the introns are removed via splicing. Control of RNA StabilityTwo protective "caps" are added to the mRNA before it leaves the nucleus to stop the strand's end from deteriorating as it travels. On the 5′ end of the mRNA, a guanosine triphosphate (GTP) molecule called the 5′ cap is often present. A string of adenine nucleotides typically makes up the poly-A tail, which is joined to the 3′ end. The amount of time that the RNA stays in the cytoplasm may be regulated once it has been delivered there. Each RNA molecule has a set lifetime and degrades at a predetermined pace. The amount of protein in the cell may be affected by this rate of degradation. The time it takes for translation to take place will be shortened if the decay rate is raised since the RNA won't be in the cytoplasm for as long. On the other side, if the rate of decay is reduced, the RNA molecule will remain in the cytoplasm for a longer period of time, allowing for the translation of more protein. This rate of disintegration is referred to as RNA stability. If the RNA is resilient, it will be visible in the cytoplasm for a longer period of time. The stability of the RNA may be impacted by protein binding. RNA-binding proteins, or RBPs, may bind in the RNA immediately upstream or downstream of the protein-coding region. Untranslated regions, or UTRs, are those parts of RNA that are not converted into protein. They are not introns since the nucleus has deleted them. Instead, these are the areas that control the translation, stability, and localization of mRNA. The 5′ UTR is the area immediately before the protein-coding region, while the 3′ UTR is the area following the coding region. The binding of RBPs to these regions may increase or decrease an RNA molecule's stability depending on the specific RBP that binds. RNA Stability and microRNAIn addition to RBPs that bind to the RNA molecule and control (increase or decrease) RNA stability, other elements known as microRNAs may adhere to it. These small RNA molecules, known as microRNAs or miRNAs, have just 21-24 nucleotides in length. The longer pre-miRNAs are converted into miRNAs in the nucleus. By use of a protein known as dicer, these pre-miRNAs are converted into mature miRNAs. Mature miRNAs recognize a particular sequence and bind to the RNA, much like transcription factors and RBPs. However, RNA-induced silencing complex (RISC), a ribonucleoprotein complex, and miRNAs also work together. RISC interacts with the miRNA to degrade the target mRNA. The RNA molecule is rapidly broken down by the RISC complex and miRNAs.
Next TopicWeight of the Human Brain
|
 For Videos Join Our Youtube Channel: Join Now
For Videos Join Our Youtube Channel: Join Now
Feedback
- Send your Feedback to [email protected]
Help Others, Please Share









