Split GenesThe split gene includes both coding (exons) as well as non-coding (introns) segments. The removal of intronic regions to form mature mRNA is called splicing. It contains at least one intron separating two or more exonic segments, while an unsplit gene has uninterrupted exonic sequences. A simple split gene must have at least two exon segments separated by one intron segment. Introns were once considered unnecessary in genes, but they actually help regulate alternative splicing patterns and RNA editing. Moreover, split genes may appear more complicated than unsplit ones because they contain non-coding regions. Still, this complexity provides more flexibility and diversity in genetic expression through mechanisms like alternative splicing and regulation by miRNAs. 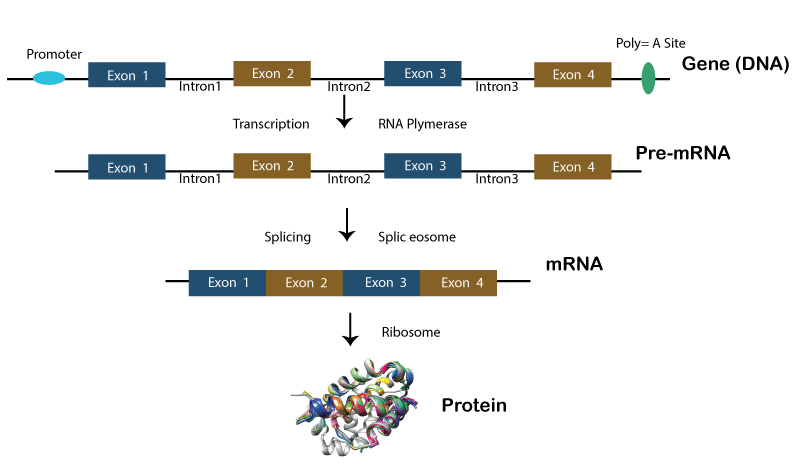
Definition of Split gene"A gene that makes pre-mRNA with introns obstructing exons is known as split genes." "A split gene contains both exons and introns. During post-transcriptional modification, non-coding introns are removed and only the protein-coding exons remain in mature RNA". Another definition of the split gene is "Whose entire sequence is not retained in RNA." Discovery of Split GenesAmerican molecular biologist Phillip Allen Sharp won the Nobel Prize in Physiology and Medicine for finding "split genes" in 1993. He discovered that split genes involving humans are the most ordinary gene structure in more elevated organisms. Richard John Roberts (1943-) also discovered split genes and shared the prize with Sharp. This discovery has been fundamental to basic research in biology and medical research on cancer and other diseases' development. Thanks to this discovery, scientists predicted the genetic process of splicing. 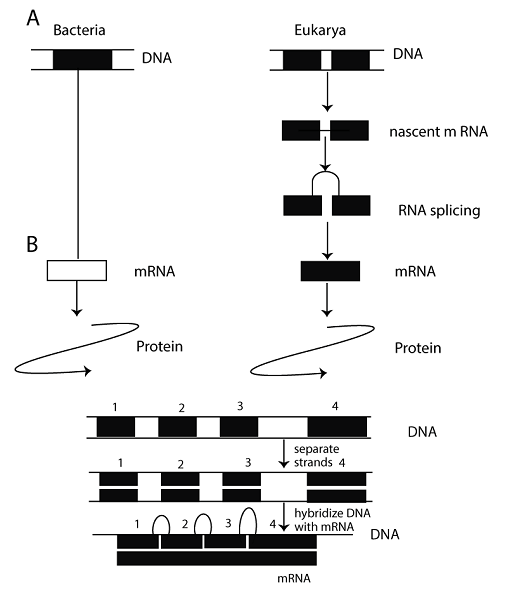
Intron, Exon Structure of Split GenesElectron micrographs of RNA-DNA hybrids and nucleotide sequencing of cloned genomic DNAs and cDNAs demonstrated that the mouse β-globin gene that encodes the β subunit of haemoglobin includes 2 introns. Moreover, these introns are spliced out from mRNA during transcription. In general, eukaryotic genes have a complicated intron-exon structure with more DNA in the introns than in the exons. For instance, the chicken ovalbumin gene has seven introns and eight exons spread over 7.7 kilobases (or 7700 base pairs) of genomic DNA. The exons add up to only about 1.9 kb, which means that roughly 75% of the gene is made up of introns. A noteworthy example is the human gene that encodes factor VIII, a blood clotting protein. This gene consists of 26 exons and approximately 186 kb of DNA. The mRNA is just almost 9 kb prolonged, revealing that introns produce up to approximately 175 kb of the gene's size. Introns normally include around 10 times more additional DNA than exons in more elevated eukaryotic genes. Many eukaryotic genes include introns, despite not all. Histone genes lack introns, demonstrating that introns are unnecessary for gene function in eukaryotes. Yeast's simple eukaryotic genes mostly lack introns. Prokaryotes have some rare genes with introns. Numerous introns don't contain a function, but few encode functional RNAs and proteins. Introns are believed to be remnants of important sequences from earlier evolution that helped accelerate it by facilitating recombination between exons of different genes - this process is called exon shuffling. Recombining introns from different genes could create new genes with unique exon combinations. Studies have shown that some genes are chimaeras of exons from multiple other genes, supporting this hypothesis. This suggests that intron sequences can facilitate the formation of novel genes through recombination. Gene Splicing MechanismGene splicing events occur after mRNA is formed from transcription, and there are several types of these events that can take place in genes. 1. Exon Skipping This Skipping is the most common gene splicing mechanism. It contains or bans exons from the last gene transcript, causing prolonged or compressed mRNA variants. Exons are responsible for producing proteins used in various cell types and functions. 2. Intron Retention In this introns are retained in the final transcript. Almost 2-5% of human genes have been notified to contain introns. The gene splicing mechanism keeps non-coding portions of the gene, resulting in abnormalities in protein structure and function. 3. Alternative 3' and 5' Splice Sites Alternative gene splicing involves joining different 5' and 3' splice sites. In this type of gene splicing, multiple alternative 5' splice sites compete to join with multiple alternative 3' splice sites. Splice Variant Detection MethodsGene splicing creates different proteins that affect human health. The best ways to detect splice variants are computational prediction and microarray analysis, with the latter being the most popular due to its sensitivity and ability to monitor gene expression on a tissue-specific, genome-wide level. Microarrays provide a reliable platform for discovering alternative gene splicing. They have detected new gene transcripts not found by other methods like ESTs or RT-PCR sequencing, which is only useful for analysing small numbers of genes. However, this method has limitations since it's labour-intensive and doesn't scale well when studying thousands of genes or hundreds of tissues. Example of Split GenesA significant instance is the human gene that encodes factor VIII, a blood clotting protein. This gene has 26 exons and approximately 186 kb of DNA. The mRNA is only about 9 kb long, meaning roughly 175 kb of the gene consists of introns. Typically, introns contain around ten times more DNA than exons in higher eukaryotic genes. The Significance of Split GenesThe role of split genes in the nucleus is not well understood, but there is significant knowledge about organelles, particularly mitochondria. The regulatory structure required for gene expression involves split genes and the nuclear envelope as missing links that permit the reconciliation of preformationism and epigenesis. A recent discovery related to intron structure and catalytic properties might create a new understanding of nucleic acid functions. Moreover, it also increases the need for reflection about intron structure and function. Architectural Limits on Split GenesExon/intron structure differs among eukaryotes, with vertebrates having large introns and small exons, while lower eukaryotes have the opposite. To study how exon and intron size affects pre-mRNA processing, internally expanded exons were added to genes in vertebrates with both small and large introns. The splicing outcome was influenced by both exon and intron size. Intron size determined if large exons were recognized efficiently; when introns were large, they skipped over the large exons but included them when they were small. Therefore, only flanking a large exon with other big introns made it incompatible with splicing. Both exon and intron sizes became problematic at about 500 nt, although their sequence also affected the recognition failure of either one. These findings suggest that gene architecture today reflects limitations on recognizing exonic sequences partly due to the pairing of splice sites during pre-mRNA recognition as well as consensus sequences aided in pairing across either an intron or an exon. ConclusionIn conclusion, Sharp and Roberts' discovery of split genes revolutionised molecular biology. It provided insights into how higher organisms, including humans, process and regulate genetic information. Understanding the structure and function of split genes also advanced medical research on cancer and other diseases. Today, scientists continue to build upon this knowledge to unravel genetic mysteries and improve human health outcomes. FAQs1. Do all eukaryotic genes have intron sequences interrupting them? No, not all eukaryotic genes are split by introns. Some genes only contain coding regions (exons), while others have both introns and exons. 2. What do split genes characterise? Split genes are only observed in eukaryotes, while no split genes have been reported in prokaryotes. Unlike eukaryotic genes, which contain segments of expressed DNA (exons) interrupted by unexpressed segments (introns), prokaryotic genes are contiguous. 3. Is the split gene an advanced feature of the genome? The split-gene arrangements are likely an ancient feature of the genome. The presence of introns is a relic of the past, and splicing represents its dominance. 4. What complications arise from split gene arrangements? Exons are sequences that appear in mature or processed RNA, interrupted by introns. Introns, or intervening sequences, do not appear in mature or processed RNA. The split-gene arrangement complicates the definition of a gene as a DNA segment. 5. Do genes divide equally? Only half of a parent's genes are passed on to each child, and siblings (except identical twins) do not inherit the exact same half. This means your siblings received some genes you did not, and vice versa. You and your siblings share about 50% of your DNA with each other. 6. When can genes be split? Linked genes can separate through recombination, which happens when two homologous chromosomes align during meiosis and exchange genetic material segments.
Next TopicAgrobacterium Mediated Gene Transfer
|
 For Videos Join Our Youtube Channel: Join Now
For Videos Join Our Youtube Channel: Join Now
Feedback
- Send your Feedback to [email protected]
Help Others, Please Share









