Protein DefinitionProteins are large biological molecules and macromolecules comprised of one or more long chains of amino acid residues. Proteins play a variety of roles in living things, including catalyzing metabolic reactions, reproducing DNA, responding to external stimuli, offering cells and organisms structure, and transporting chemicals. The primary way that proteins vary from one another is by the arrangement of their amino acids, which is governed by the nucleotide sequence of their genes. This sequence causes the protein to typically fold into a particular 3D structure, which controls its activity. A polypeptide is an ordered sequence of amino acid residues. At least one lengthy polypeptide is present in every protein. Polypeptides of 20-30 residues or fewer are frequently referred to as peptides rather than proteins and sometimes also called short polypeptides. Peptide bonds and nearby amino acid residues hold the residue of the individual amino acid together. The arrangement of amino acid residues in a protein is determined by the sequence of a gene, which is encoded in the genetic code. 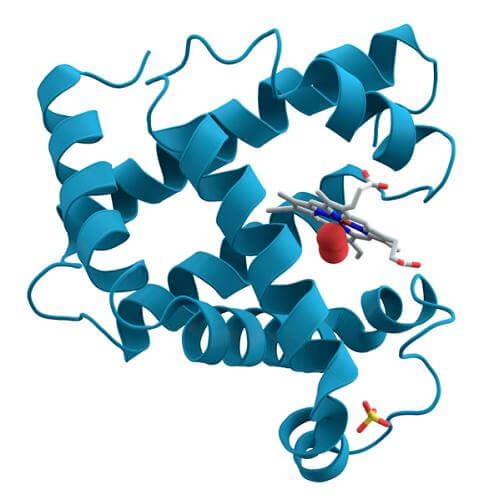
The genetic code typically only defines the 20 standard amino acids, but in some organisms, it may also include selenocysteine and-in some archaea-pyrrolysine. Just after or even throughout protein synthesis, the residues are regularly chemically changed through post-translational modification. As a result, the physical and chemical properties, activity, folding, stability, and ultimately the functioning of the proteins are affected. Non-peptide groups, often known as cofactors or prosthetic groups, are sometimes added to proteins. Additionally, proteins can cooperate in carrying out certain tasks, and they frequently join forces to create stable protein complexes. Protein turnover is the process through which the machinery of the cell breaks down and recycles proteins that have already been created after a finite amount of time. The half-life of a protein is a broad measure of a protein's lifespan. In mammalian cells, they typically live for 1-2 days, though they might last for minutes or years. Proteins that are abnormal or misfolded degrade more quickly either because they are targets for apoptosis or because they are unstable. Proteins play a crucial role in nearly every cellular process and are fundamental components of organisms, just like other biological macromolecules like polysaccharides and nucleic acids. Enzymes are common in proteins and are vital for metabolism and the catalysis of biological processes. Proteins that also have structural or mechanical purposes include actin and myosin, which are found in muscle, as well as the proteins that make up the cytoskeleton, which forms a scaffolding framework that maintains cell shape. Origin and historyAntoine Fourcroy and his fellow mates recognized proteins as a separate class of biomolecules in the eighteenth century. These molecules were characterized by their tendency to coagulate or flocculate in the presence of heat or acid. Blood serum albumin, albumin from egg whites, fibrin, and wheat gluten were prominent examples at the time. The Swedish scientist Jöns Jacob Berzelius and the Dutch chemist Gerardus Johannes Mulder both originally identified proteins in 1838. Common proteins were subjected to elemental analysis by Mulder, who discovered that almost all proteins shared the same empirical formula, C400H620N100O120P1S1. He incorrectly assumed that they might be made up of a single kind of (extremely huge) molecule. These molecules were referred to as "proteins" by Mulder's colleague Berzelius; the word "protein" comes from the Greek word proteios, which means "principal", "in the lead," or "standing at the front," + -in. Mulder continued by identifying the byproducts of protein cleavage, including the amino acid leucine, for which he discovered a (roughly accurate) molecular weight of 131 Da. Before "protein," other terms like "albumins" or "albuminous materials" were employed. Since "flesh makes flesh," early nutritional researchers like the German Carl von Voit considered that protein constituted the most crucial nutrient for preserving the body's structure. Karl Heinrich Ritthausen discovered glutamic acid and expanded the range of known protein forms. Thomas Burr Osborne at the Connecticut Agricultural Research Institute wrote a thorough analysis of vegetable proteins. The amino acids that are necessary for nutrition were found through collaboration with Lafayette Mendel and the use of Liebig's law of the lowest in nourishing laboratory rats. William Cumming Rose maintained the project and disseminated the information. Franz Hofmeister and Hermann Emil Fischer's research led to the ability to visualize proteins as polypeptides in 1902. It wasn't until James B. Sumner demonstrated that the enzyme urease was actually a protein in 1926 that the crucial significance of proteins as enzymes in living organisms was clearly understood. Proteins were extremely challenging for early protein biochemists to research because they were so difficult to purify in significant numbers. Early research, therefore, concentrated on purifiable proteins in high numbers, such as those found in blood, egg white, different poisons, and digestive/metabolic enzymes collected from slaughterhouses. The Armour Hot Dog Co., in the 1950s, distilled 1 kg of pure bovine pancreatic ribonuclease A and made it available to researchers for free; this move aided ribonuclease A's development into an important topic of biochemical research in the subsequent decades. Linus Pauling is credited for developing William Astbury's successful hydrogen bonding-based prediction of regular protein secondary structures in 1933. Walter Kauzmann's later work on denaturation, which was partially based on earlier findings by Kaj Linderstrm-Lang, added to our knowledge of protein folding and structure mediated by hydrophobic interactions. In 1949, Frederick Sanger sequenced the first protein, insulin. By properly identifying the amino acid composition of insulin, Sanger proved beyond a shadow of a doubt whether proteins were made of linear polymers of amino acids instead of branching chains, colloidal matter, or cyclols. For this accomplishment, he received the Nobel Prize in 1958. X-ray crystallography's advancement made it possible to sequence protein structures. In 1958, John Kendrew and Max Perutz, respectively, established the very first protein complexes for myoglobin and hemoglobin. The sequencing of complicated proteins was made possible by computer use and rising computational power. Roger Kornberg used high-intensity X-rays from synchrotrons in 1999 to successfully sequence the extremely complicated structure of RNA polymerase. Since then, huge macromolecular assembly cryo-electron microscopy (cryo-EM) has been created. Instead of crystals, cryo-EM analyses protein samples that have been frozen, and it does so using electron beams rather than x-rays. It results in less harm to the sample, enabling researchers to get more data and examine more complex structures. Researchers have also made progress toward atomic-level resolution of protein structures thanks to computational protein structure prediction of tiny protein domains. In the Protein Data Bank, as of 2017, there were over 126,060 atomic-resolution protein structures. How many Proteins are Encoded in Genomes?Approximately equal numbers of proteins and genes are found in each genome (although there may be a significant number of genes that encode RNA of protein, e.g., ribosomal RNAs). Eukaryotes frequently encode tens of thousands of proteins, in comparison to bacteria and archaea, which typically synthesize a few hundred to a few thousand proteins. BiochemistryMost proteins are linear polymers made up of up to 20 distinct L-amino acid sequences. All proteinogenic amino acids share the same structural element, the -carbon, where a carboxyl group, an amino group, and a changeable side chain are attached. Only proline deviates from this fundamental structure because it has an unusual ring at the N-end of the amine group, which locks the CO-NH amide moiety into a locked conformation. A protein's three-dimensional structure and chemical reactivity are ultimately defined by the combined influence of all of its amino acid side chains, which are listed in the standard list of amino acids and have a variety of chemical structures and properties. 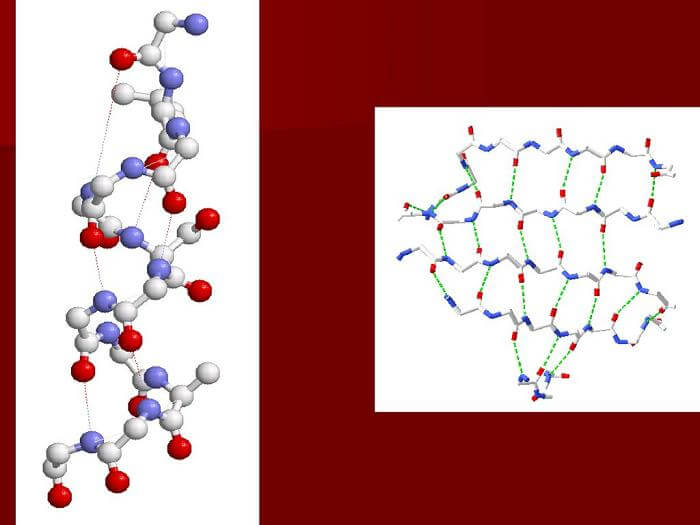
In a polypeptide chain, an individual amino acid is known as a residue once it has been linked to another amino acid in a protein chain. The main chain, also known as the protein backbone, is made up of linked groups of carbon, nitrogen, and oxygen atoms. Two resonance forms of the peptide bond, which give certain double-bond characteristics and prevent rotation along its axis, help to maintain the alpha carbons' approximate coplanarity. The local form that the protein backbone takes is governed by the second and third dihedral angles, which are present in the peptide bond. The free amino group is found at the protein's N-terminus, also known as the amino terminus, while the free carboxyl group is found at the protein's C-terminus, also known as the carboxy terminus. It might be difficult to distinguish between the meanings of the phrases protein, polypeptide, and peptide. The term "protein" typically refers to the entire biological molecule in a stable shape, while the term "peptide" is typically reserved for short amino acid oligomers that frequently lack a stable 3D structure. But the line between the two is not clearly defined, and it typically lies between 20 and 30 residues. A chain of amino acids, which is linear, can be referred to as a "polypeptide," normally irrespective of length. However, this phrase frequently implies the absence of a particular structure. InteractionsProteins can interact with a wide variety of substances, including DNA, lipids, carbohydrates, and other proteins. Cells' AbundanceBacteria of average size are thought to possess roughly 2 million proteins per cell (e.g., Staphylococcus aureus and E. coli). The number of molecules in smaller bacteria, including spirochetes and Mycoplasma, ranges from 50,000 to 1 million. Eukaryotic cells, on the other hand, are bigger and have a lot more protein within. For instance, it is estimated that human cells contain between 1 and 3 billion and 50 million proteins, respectively, in yeast cells. A few molecules to 20 million copies of a single protein can be found in a given cell. Most cells do not express every gene that codes for a protein, and the number of expressed genes varies depending on factors like cell type and environmental stimulation. Just 6,000 of the roughly 20,000 proteins which are encoded by the human genome, for instance, are present in lymphoblastoid cells. SynthesisUsing the instructions stored in genes, amino acids are combined to form proteins. The gene that encodes for each protein and its nucleotide sequence determines the exact amino acid sequence of that protein. Each three-nucleotide combination in the genetic code, known as codons, designates an amino acid. For instance, the code for methionine is AUG (adenine-uracil-guanine). 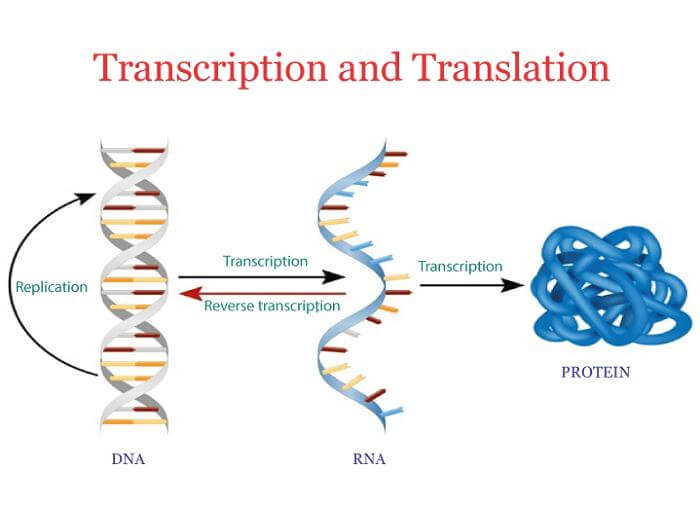
There is some redundancy in the genetic code because there are 64 possible codons because DNA has four nucleotides. As a result, some amino acids are specified by more than one codon. Pre-messenger RNA (mRNA) is created by the transcription of genes from DNA into proteins like RNA polymerase. The majority of organisms process the pre-mRNA (also referred to as a main transcript) through various post-transcriptional modifying processes to produce the mature mRNA. The ribosome then uses the mature mRNA as a template for protein synthesis. Prokaryotes either utilize their mRNA immediately after they make it or bind it after it has traveled out from the nucleoid. In contrast, mRNA is created in the cell nucleus of eukaryotes and then transported over the nuclear membrane further into the cytoplasm, where it is used to synthesize proteins. In comparison to eukaryotes, prokaryotes may synthesize proteins at a rate of up to 20 amino acids in a second. A protein is made from an mRNA template through a process known as translation. The mRNA is loaded onto the ribosome and read three nucleotides at a time by matching each codon to its base pairing anticodon on a transfer RNA molecule, which carries the amino acid necessary for the codon it recognizes. The necessary amino acids are "charged" onto the tRNA molecules by aminoacyl tRNA synthetase. The term "nascent chain" frequently refers to the expanding polypeptide. From the N-terminus to the C-terminus, proteins are always biosynthesized. Chemical SynthesisShort proteins can also be made chemically using the peptide synthesis method, which relies on methods for high-yield peptide synthesis, including chemical ligation. Polypeptide chains can contain non-natural amino acids thanks to chemical synthesis, which also enables the attachment of fluorescent probes to the side chains of amino acids. 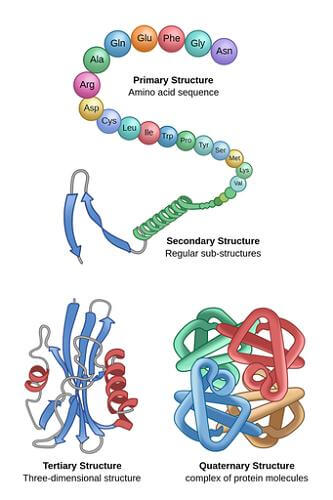
Even though they are typically not used for commercial applications, these techniques are helpful in laboratory biochemistry and cell biology. For polypeptides longer than 300 amino acids, chemical synthesis is ineffective, and the produced proteins might not quickly adopt their natural tertiary structure. Contrary to a biological response, the majority of chemical synthesis processes go from C-terminus to N-terminus. Proteins typically fold into distinctive 3D shapes. The native conformation of a protein refers to how it naturally folds. Although many proteins can fold freely due to the chemical properties of their amino acids, others, to fold into their native states, require the assistance of molecular chaperones. Four distinctive features of a protein's structure are frequently mentioned by biochemists: The amino acid sequence is the basic structure. A polyamide is a protein. The secondary structure is composed of locally repeated patterns that are supported by hydrogen bonds. The three most prevalent examples are the -helix, -sheet, and twists. Numerous regions of various secondary structures can exist in the same protein molecule because secondary structures are localized. The tertiary structure is composed of the overall structure of a single protein complex and the spatial configuration of the secondary structures. Tertiary structure is typically maintained through nonlocal interactions, including the formation of a hydrophobic region, salt bridges, hydrogen bonds, disulfide bonds, and even posttranslational modifications. The fold is frequently used interchangeably with the phrase "tertiary structure." The fundamental operation of the protein is under the control of the tertiary structure. The next structure is made up of many protein molecules (polypeptide chains), also known as protein subunits in this context, that work as a single protein complex and is known as a quaternary structure. Quinary structure- The distinguishing characteristics of protein surfaces that arrange the crammed cellular interior. The quinary structure depends on fleeting yet crucial macromolecular interactions that take place inside live cells. Proteins are not completely rigid molecules. Along with these layers of structure, proteins can switch between a variety of related structures as they carry out their jobs. When discussing these functional translocations, these tertiary or quaternary forms are sometimes described as "conformations," and changes between them are known as conformational shifts. Such alterations often follow the interaction of a reactant molecule to the active site of an enzyme or the region of the protein engaged in chemical catalysis. Proteins' structural variation in solution is also influenced by thermal vibration and molecular collisions. The three basic classes of proteins-globular, fibrous, and membrane-that correspond to typical tertiary structures can be arbitrarily grouped into three groups. Numerous globular proteins are enzymes, and almost all of them are soluble. Like collagen, the main protein in connective tissue, or keratin, the protein in hair and nails, fibrous proteins are frequently structural. A lot of times, membrane proteins act as receptors or as passageways for polar or charged molecules to flow across the cell membrane. Dehydrons are a specific type of protein that exhibits intramolecular hydrogen bonds that are inadequately protected against water attack and so encourage their own dehydration. Regions of ProteinsProteins are made up of many protein domains, or pieces of a protein that wrap into various structural units. In most cases, domains also serve specialized purposes, such as enzymatic activity (such as kinase) or acting as binding modules (e.g., Other proteins with proline-rich sequences can bind to the SH3 domain.). Series MotifProteins frequently serve as recognition sites for other proteins by including short amino acid sequences. As an illustration, SH3 domains frequently bind to short PxxP motifs (i.e., Divided by two unknown amino acids [x], there are two prolines [P], albeit the neighboring amino acids could affect the precise binding affinity). The Eukaryotic Linear Motif (ELM) database contains many of these motifs. Cellular ProcessesThe primary actors in a cell are proteins, which are thought to carry out the functions dictated by the data contained in genes. The majority of other biological molecules are relatively innocuous substrates for proteins, with the exception of some forms of RNA. While other macromolecules like DNA and RNA compensate only 3% and 20%, respectively, of the dry weight of an Escherichia coli cell, proteins constitute up half of it. A cell or type of cell's proteome is the collection of proteins that are expressed in those cells. 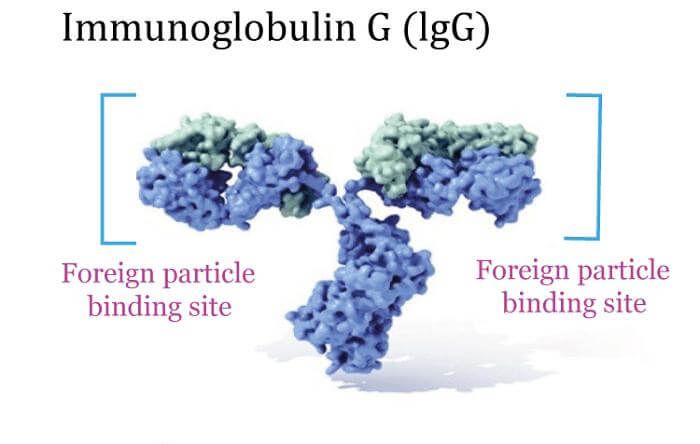
Proteins' main property, which also enables their wide range of tasks, is their capacity to bind other molecules firmly and specifically. The binding site, which is a depression or "pocket" on the molecular surface, is the area of the protein that is in charge of attaching to another molecule. The side chains of the surrounding amino acids and the tertiary structure of the protein, which creates the ligand binding region, both contribute to moderating this binding capacity. The ribonuclease receptor protein, for example, binds to human angiogenin with a sub-femtomolar dissociation constant (1015 M) but not to its amphibian homolog increase (>1 M), despite having a higher dissociation constant. This is an example of how protein binding can be extremely selective and tight. When a single methyl group is added to a binding partner, for instance, binding can sometimes be almost completely eliminated. For instance, the very identical side chain of the amino acid isoleucine is discriminated against by the aminoacyl tRNA synthetase that is specific to the amino acid valine. Both small-molecule substrates and other proteins can bind to proteins. Proteins can oligomerize to create fibrils when they specifically bind to other copies of the same molecule; this is a process that frequently happens in structural proteins, which are made up of globular monomers that self-associate to form hard fibers. Protein-protein interactions drive enzymatic activity, direct cell cycle development, and enable the formation of massive protein complexes that perform a variety of closely linked biological functions. Proteins can adhere to cell membranes or even incorporate themselves into them. Protein conformational changes brought about by binding partners enable the creation of incredibly intricate signaling networks. Since protein interactions are reversible and heavily rely on the presence of various partner protein groups to form aggregates that can carry out distinct sets of functions, studying the interactions between particular proteins is essential to understanding key facets of cellular function and, ultimately, the characteristics that set different cell types apart. EnzymesProteins' most well-known function in a cell is as enzymes, which speed up chemical processes. Enzymes often accelerate just one or a few chemical reactions and are quite specialized. The majority of metabolic reactions are carried out by enzymes, which also manipulate DNA through procedures including transcription, DNA repair, and replication. In a process called posttranslational modification, some enzymes modify other proteins by adding or removing chemical groups. Enzymes are known to catalyze about 4,000 processes. Large rate accelerations are frequently the outcome of enzyme catalysis; for example, orotate decarboxylase accelerates the rate by 1017 times compared to the uncatalyzed process. Substrates are the molecules that enzymes bind to and react with. Though enzymes can include hundreds of amino acids, typically only a fraction of the residues-three to four residues on average-are directly engaged in catalysis, and only a very small portion of the total residues come into contact with the substrate. The term "active site" refers to the area of an enzyme that binds the substrate and houses the catalytic residues. Members of the protein class known as divergent proteins control the stereochemistry of a molecule produced by another enzyme. Ligand Binding and Cell SignalingSignal transduction and cell signaling are processes that involve a large number of proteins. Some proteins, like insulin, are intercellular and communicate with other cells in other areas from the cell in which they were created. Others are receptor membrane proteins whose primary job is to bind a signaling molecule and cause a biological reaction in the cell. The internal effector domain of many receptors can be active enzymatically or undergo a conformational change that can be recognized by other proteins. Many receptors also have a disclosed binding site on the cell surface. Antibodies, protein-based components of the adaptive immune system, are primarily responsible for binding antigens or foreign substances in the body and directing the immune system's eradication process toward them. Antibodies can either be released into the extracellular environment, or they can be released and attached to the membranes of plasma cells, which are specialized B cells. Enzymes are constrained in their ability to bind to their substrates by the requirement to carry out their reaction, but antibodies are not subject to these restrictions. An antibody's binding affinity to its target is quite high. Numerous ligand transport proteins bind specific tiny biomolecules and move them to different parts of a multicellular organism's body. An antibody's binding affinity to its target is quite high. These proteins need to have a high binding affinity when the ligand is available in small levels in the target tissues and a low binding affinity when it is present in large numbers. Haemoglobin, which transports oxygen through the lungs to various tissues and organs in all vertebrates and has close homologs in each and every biological kingdom, is the most well-known example of a ligand-binding protein. Lectins are proteins that bind sugar and are incredibly selective for the sugar moieties they recognize. Biological recognition processes involving cells and proteins frequently involve lectins. Hormones and receptors are extremely selective binding proteins. The permeability of the cell membrane to ions and small molecules can be altered by transmembrane proteins acting as ligand transport proteins. Polar or charged molecules cannot diffuse through the membrane's hydrophobic core on their own. Internal channels in membrane proteins allow these chemicals to enter and leave the cell. Many ion channel proteins are tailored to discriminate for just one specific ion; for instance, potassium and sodium channels frequently only do so for one of the two ions. Building Block ProteinsIn contrast to fluid biological components, structural proteins give them firmness and rigidity. The bulk of structural proteins is fibrous proteins. For example, keratin is a constituent of hard or filamentous structures like hair, nails, feathers, hooves, and some animal shells, whereas collagen and elastin are crucial components of connective tissue like cartilage. Some globular proteins can also serve structural purposes. For instance, actin and tubulin are globular and soluble as monomers, but when they polymerize, they create long, stiff fibers that make up the cytoskeleton and help the cell maintain its size and shape. Motor proteins like myosin, kinesin, and dynein, which are able to produce mechanical forces, are other proteins that have structural roles. Both the sperm of many multicellular animals that engage in sexual reproduction and the cellular motility of single-celled species depend on these proteins. Evolution of ProteinsHow proteins evolve, or more specifically, how many mutations (or rather changes in amino acid sequence) result in new structures and functions, is a central subject in molecular biology. The majority of amino acids in a protein can be changed without compromising activity or function, as evidenced by the abundance of homologous proteins found in other species (as compiled in specialized databases for protein sequences, e.g., PFAM). A gene may be replicated before it can mutate naturally to avoid the dramatic effects of mutations. Pseudogenes can result from this as well as the full loss of gene activity. Although some can significantly alter protein function, particularly in enzymes, most single amino acid alterations have only minor effects. For instance, a single mutation or a few mutations might alter the substrate specificity of several enzymes. An enzyme's selectivity and, subsequently, its enzymatic activity can change as a result of mutations. Bacteria (or other organisms) can therefore adapt to many food sources, even synthetic substrates like plastic. NutritionAnimals (including humans) and plants can typically biosynthesize all 20 of the essential amino acids, although most bacteria and plants cannot. Known as essential amino acids, these are the amino acids that a living thing cannot produce on its own. Animals are deficient in several critical enzymes, including aspartokinase, which catalyzes the initial stage of the production of lysine, methionine, and threonine from aspartate. If the environment contains amino acids, bacteria can save energy by consuming the amino acids from the environment and reducing the activity of their biosynthetic processes. 
Animals receive amino acids through the intake of protein-rich meals. Proteins that have been consumed are then broken down into amino acids during digestion, which normally entails denaturing the protein by exposing it to acid and hydrolyzing it by protease enzymes. While some ingested amino acids are being used for protein production, others are converted to glucose by gluconeogenesis or supplied into the citric acid cycle. Under times of malnutrition, the body's own proteins, especially those located in muscle, can be used to keep us alive thanks to this utilization of protein as fuel. By encouraging hair follicle growth and keratinization in animals like dogs and cats, protein preserves the health and quality of the skin and lowers the risk that skin conditions may cause odors. Poor-quality proteins can also affect digestive health, raising the risk of flatulence and odorous substances in dogs because when proteins enter the colon undigested, they are fermented, resulting in the production of hydrogen sulfide gas, indole, and skatole. Animal-based proteins are more easily absorbed by dogs and cats than those derived from plants, but inferior animal products, including skin, feathers, and connective tissue, are indigestible. ConclusionHuman body is made up of millions of cells and every singlebin a human body consists of protein. The most basic and simplified structure of a protein is a chain of amino acids. Protein is very essential for survival of life on this earth. Our diet must contain protein in order to repair the demaged cells also make new ones. It also plays a vital role in growth and development of every organism including humans.
Next TopicSustainable Development Definition
|
 For Videos Join Our Youtube Channel: Join Now
For Videos Join Our Youtube Channel: Join Now
Feedback
- Send your Feedback to [email protected]
Help Others, Please Share









